Abstract
It is a worthy goal to completely characterize all human proteins in terms of their domains. Here, using the Pfam database, we asked how far we have progressed in this endeavour. Ninety per cent of proteins in the human proteome matched at least one of 5494 manually curated Pfam-A families. In contrast, human residue coverage by Pfam-A families was <45%, with 9418 automatically generated Pfam-B families adding a further 10%. Even after excluding predicted signal peptide regions and short regions (<50 consecutive residues) unlikely to harbour new families, for ∼38% of the human protein residues, there was no information in Pfam about conservation and evolutionary relationship with other protein regions. This uncovered portion of the human proteome was found to be distributed over almost 25 000 distinct protein regions. Comparison with proteins in the UniProtKB database suggested that the human regions that exhibited similarity to thousands of other sequences were often either divergent elements or N- or C-terminal extensions of existing families. Thirty-four per cent of regions, on the other hand, matched fewer than 100 sequences in UniProtKB. Most of these did not appear to share any relationship with existing Pfam-A families, suggesting that thousands of new families would need to be generated to cover them. Also, these latter regions were particularly rich in amino acid compositional bias such as the one associated with intrinsic disorder. This could represent a significant obstacle toward their inclusion into new Pfam families. Based on these observations, a major focus for increasing Pfam coverage of the human proteome will be to improve the definition of existing families. New families will also be built, prioritizing those that have been experimentally functionally characterized.
Database URL: http://pfam.sanger.ac.uk/
Introduction
The sequencing of the human genome (1) and large-scale projects such as ENCODE (2) have provided access to a more complete and reliable list of human protein-coding genes than was previously available. The current collection of human proteins that are available from the manually reviewed UniProtKB/Swiss-Prot database (3) is just over 20 000 sequences. This list, while still being updated, has become more stable in recent times. Full functional characterization of this set of proteins is expected to deliver a finer understanding of how human cells develop, function and interact.
Pfam (4) is a collection of families composed of homologous protein regions. There are two distinct sets of Pfam families: a manually curated collection called Pfam-A and an automatically generated set termed Pfam-B. Starting from a seed alignment of homologues, the profile hidden Markov model (HMM)-based package HMMER3 (http://hmmer.janelia.org/) is used to build a representative model for a Pfam-A family that is then run against the UniProtKB database (3) to detect more homologous family members. Each Pfam-A family is functionally annotated by a curator using information from the literature, when available. The Pfam-B set of families consists of automatically generated unannotated regions of sequence conservation that are not currently represented by a Pfam-A entry. The Pfam-B alignments are initiated from the clusters contained in the ADDA database, which are generated from clustering a 40% non-redundant version of UniProtKB (5, 6). Pfam release 27.0 contains 14 831 Pfam-A families and 544 963 Pfam-B families.
Pfam and other databases that group proteins into families can contribute to functional characterization of the human proteome. They detect conserved functional modules, typically sub-sequences, which link human protein regions to their homologues within human and across other species. Identification of these links can generate functional hypotheses via homology-based annotation transfer, even in cases when sequence conservation does not span the full length of the proteins involved. For example, it can highlight that the sequence similarity between the UniProtKB sequences P62993 (growth factor receptor-bound protein 2; Grb2) and P12931 (tyrosine-protein kinase Src) is located in the SH2 (PF00017) and SH3 (PF00018) Pfam-A domains, two commonly occurring protein-binding modules (7, 8), rather than reflecting any shared enzymatic function; Grb2 is not known to have enzymatic action (9). Identification and annotation of homologous regions can also help comparative genomics and reconstruction of the evolutionary history of proteins.
Here, we ask how much of the human proteome is currently covered by the conserved regions that constitute Pfam families and what challenges lie ahead in achieving our goal of a more complete annotation of similar regions.
Methods
Human, Saccharomyces cerevisiae and Escherichia coli proteomes
We downloaded the UniProtKB/Swiss-Prot-reviewed protein sequences for Homo sapiens (taxonomic identifier 9606; 20 234 sequences), S. cerevisiae strain ATCC 204508 /S288c (taxonomic identifier 559292; 6621 sequences) and E. coli strain K12 (taxonomic identifier 83333; 4431 sequences) from the UniProt website (http://www.uniprot.org/) (3). The specific strains of E. coli and S. cerevisiae downloaded were chosen as they have the most complete protein set for these organisms in UniProtKB/Swiss-Prot (personal communication with the UniProt team).
Pfam-A and Pfam-B assignments
The human, S. cerevisiae and E. coli proteomes were searched against the Pfam-A families from Pfam 27.0, with the Pfam curated bit score gathering thresholds used to decide significant matches. We extracted the Pfam-B families for the human proteins from Pfam 27.0.
Sequence and amino acid coverage of the human proteome
Sequence coverage is defined as the percentage of sequences in a given set (e.g. the human proteome) that has a match to at least one Pfam family. The sequence is counted as covered even if the Pfam match or matches align to only part of it. Amino acid coverage for the same sequence set is defined as the percentage of residues in that set that can be aligned to Pfam families profile HMMs (using HMMER3 envelope co-ordinates).
Investigating regions uncovered by Pfam using phmmer
We identified 24 896 human regions in the human proteome not covered by Pfam-A or Pfam-B, that are not predicted to include signal peptides and that are ≥50 amino acids. We refer to these as the ‘uncovered regions’. HMMER3 phmmer (http://hmmer.janelia.org/) searches against UniProtKB release 2012_06 were run using the uncovered regions as query and both domain and sequence E-value inclusion thresholds of 10−3. We counted the number of homologous regions retrieved from UniProtKB for each search, and calculated how many of the homologous regions (using HMMER3 alignment co-ordinates) overlapped with the alignment co-ordinates of a Pfam-A family from Pfam 27.0.
MCL clustering
The uncovered regions were clustered using the MCL suite of programs (10). As input to the program, we used the sequence E-values from an all against all phmmer search on the uncovered regions (note that no E-value higher than 10−3 was considered). For the inflation value I, or the parameter that regulates the granularity of clustering in MCL, we tried the three values suggested on the MCL website, i.e. 1.4, 2.0 and 6.0. The number of clusters for the three cases was 15 266, 15 546 and 15 904, respectively. Manual inspection of region membership of the top clusters suggested that lower granularity would be a reasonable choice for a first, if rough, estimate of the number of independent clusters. For further analysis, we hence selected the 15 266 clusters obtained using I = 1.4. For each cluster, we calculated the mean number of phmmer UniProtKB matches and Pfam-A overlaps (mean over all members in the cluster).
SUPERFAMILY and Gene3D coverage of the human proteome
We downloaded the file of UniProtKB Gene3D data from ftp://ftp.biochem.ucl.ac.uk/pub/gene3d_data/v11.0.0/uniprot_assignments.csv.gz. In all, 19 984 out of the 20 234 (99%) human proteins were successfully mapped to a sequence in the file. We calculated the sequence and residue coverage for these 19 984 human sequences. We ran the human proteome against SUPERFAMILY 1.75 using InterProScan (11) with default parameters. Note that we used a pre-released version of InterProScan, version 5RC4, which was provided to us by the InterPro team. Using the resulting SUPERFAMILY 1.75 match data, we calculated the sequence and residue coverage of the human proteome.
Prediction of compositionally biased regions
The presence of transmembrane helices, coiled-coil regions, low-complexity regions, signal peptides and disordered regions was predicted in the human proteome. Coiled-coil and low-complexity regions were predicted using default parameters with ncoils (http://www.russelllab.org/cgi-bin/coils/coils-svr.pl) and SEG (12), respectively. Transmembrane and signal peptide regions were predicted using Phobius (13) with the default options. Disordered regions were predicted using IUPred (14) with the long option.
Disordered regions using the D2P2 database
In all, 18 240 of the 20 234 (90%) human proteins were successfully mapped to a sequence in the D2P2 database (15). Disorder predictions were available for the following nine prediction methods: Espritz-D, Espritz-N, Espritz-X, IUPred-L, IUPred-S, PrDOS, PV2, VLXT, VSL2b. For each residue in the 18 240 human sequences for which D2P2 data were available, we calculated how many of the nine methods predicted the residue to be disordered, and the Pfam-A coverage of these residues. The length of the disordered region for each sequence was also calculated.
Results
Homo sapiens sequence coverage was higher than that of S. cerevisiae and close to that of E. coli
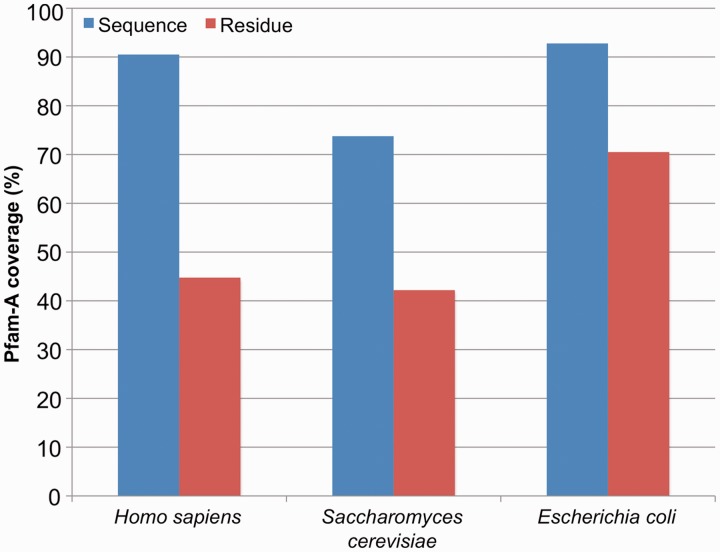
Pfam-A families from Pfam 27.0 covered 90% of proteins in H. sapiens. That is, 9 out of 10 human proteins had a match to at least one Pfam-A family. Overall, 45% of all residues in the human proteome could be assigned to a Pfam-A family. These numbers can be compared with the Pfam-A coverage for E. coli and S. cerevisiae (budding yeast), which are generally considered to be the best annotated prokaryotic and eukaryotic organisms, respectively (16, 17). The Pfam-A sequence coverage of H. sapiens was better than that of S. cerevisiae and more or less on a par with that of E. coli (Figure 1, blue bars). At the residue level, however, coverage of human was comparable with S. cerevisiae (45% and 42%, respectively) but well below E. coli coverage (70%) (Figure 1, red bars).
Figure 1.
Pfam-A coverage of H. sapiens, S. cerevisiae and E. coli. Sequence coverage (blue) is calculated as the percentage of the proteome (Methods) that matches at least one Pfam-A family. Residue coverage (red) is the percentage of amino acids in the proteome that are covered by a Pfam-A family.
Thousands of human Pfam-B families
Although it should be stressed that no direct comparison can be drawn between Pfam-B and future Pfam-A families, the former can be used as a rough estimate of the number of Pfam-A families needed to cover the same protein regions. There were 9418 Pfam-B families that matched human protein residues that were not covered by Pfam-A families. Together these Pfam-B families provided ∼10% additional residue coverage. There was no small set of Pfam-B families that could provide a substantial increase in coverage. To demonstrate this, we ranked the Pfam-B families according to amino acid coverage. The top 500 Pfam-B families with highest coverage provided <3% additional residue coverage, while the top 1000 families provided <4% additional residue coverage.
Fifteen thousand additional human clusters in search of classification
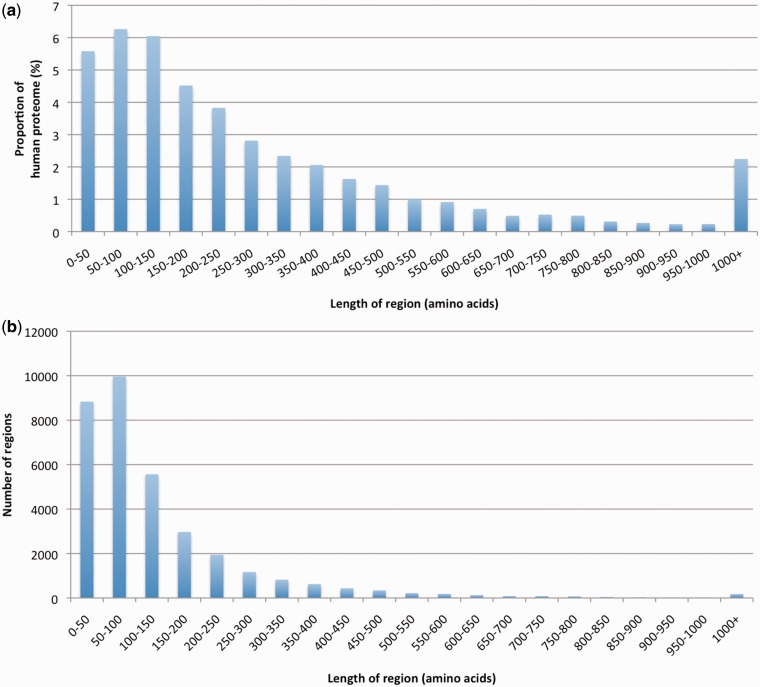
Together, Pfam-A and Pfam-B families covered ∼55% of the residues in the human proteome, leaving 45% uncovered. In Figure 2a, we plotted the length of continuous segments of amino acids that were not in Pfam-A or Pfam-B against the proportion of the human proteome that they accounted for. In Figure 2b, we showed the length distribution of these segments. We saw that uncovered regions shorter than 50 amino acids made up close to 6% of the proteome. As relatively few protein domains are shorter than 50 residues, the majority of these regions were likely to be either non-conserved linkers between already annotated families, or small extensions/variations of the flanking families. If we discounted regions of <50 residues, and regions predicted to be signal peptides (the latter representing <1% of the human proteome), we were left with ∼38% of human residues unaccounted for; these residues were found in a total of 24 896 regions. To better understand how these 24 896 regions related to each other, we ran phmmer (http://hmmer.janelia.org/) all versus all to detect similarity between them, and used the program MCL (10) to cluster them based on E-values (Methods). Using MCL inflation parameters I of 1.4, 2.0 and 6.0, we obtained 15 266, 15 546 and 15 904 clusters, respectively. Note that MCL provided just a first rough estimate of the number of unrelated clusters represented in this set of human regions. This was nonetheless a useful first step to inform the analysis discussed hereafter. In the following, we considered the clusters obtained using I= 1.4 (i.e. the parameter value giving the lowest degree of granularity).
Figure 2.
(a) Proportion of the human proteome contained in regions that are not part of Pfam-A, Pfam-B or of a predicted signal peptide versus region length. (b) Length distribution of human regions in (a).
To look at relationships between the MCL clusters and proteins in UniProtKB, we used the member regions in each cluster as queries for phmmer searches against UniProtKB (version 2012_06) applying a sequence and domain E-value inclusion threshold of 10−3 (Methods). We found that <0.1% of regions matched only human sequences in UniProtKB. For each cluster, we calculated the mean number of regions in UniProtKB that matched the cluster members and found that 9794 (64%) clusters had a mean of <100. In contrast, only 310 (2%) clusters matched ≥1000 regions in UniProtKB.
Findings from the top 10 largest clusters of human regions not in Pfam
In Table 1, we list the 10 largest MCL clusters (i.e. the ones comprising the highest number of members). We looked at these clusters in some detail to see whether they might represent interesting, yet undiscovered, new human families. Cluster 1 consisted largely of N-terminal regions in UniProtKB proteins annotated as olfactory receptors. Olfactory receptors in Pfam are grouped under family PF13853 (7tm_4; 25 558 domains in Pfam 27.0), which is part of the CL0192 (GPCR_A) clan. Inspection of family PF13853 revealed that sequences part of the seed alignment do not include the first three (N-terminal) predicted transmembrane helices of the seven that are characteristic of these receptors. We hence modified family PF13853 by extending the alignment to cover the whole predicted transmembrane domain. As a consequence, cluster 1 regions were incorporated into an updated version of the PF13853 family, which will be available in Pfam 28.0. As an additional benefit, alignment extension of PF13853 allowed us to map 6461 more domains to the family, bringing its total number of members in UniProtKB (version 2012_06) to 32 019. Most cluster 2 and 3 members were part of proteins that featured numerous zinc finger motif repeats in other parts of their sequences. Additionally, their phmmer matches in UniProtKB included a large number of regions that were already classified in Pfam as zinc fingers suggesting this as the likely classification for these regions. It has been previously observed (6) that HMMER version 3.0 has a lower sensitivity on short, very divergent families with respect to version 2.3.2. Thus, current Pfam families and clans may not be adequately covering repeats and short motifs. We are hopeful that future HMMER algorithmic developments may resolve many of the issues associated with these protein regions. The other possibility would be to try to re-organize the zinc finger clan and some of its families in order to additionally include these human regions. Similar such cases were found in clusters 4, 5, 6, 7 and 8 in which member regions often aligned to UniProtKB regions falling into the collagen repeat (PF01391), the YWTD motif repeat (PF00058), the leucine-rich repeat clan (CL0022), the EGF domain clan (CL0001) and the ‘Classical-C2H2 and C2HC zinc fingers’ clan (CL0361), respectively. Cluster 9 included regions whose UniProtKB matches were often found in the PRY (PF13765) family and hence likely to be divergent elements of that family not captured by the current PRY profile HMM. Finally, based on the domain architecture of the sequences on which the member regions occurred, cluster 10 was likely to represent a cadherin cytoplasmic domain not yet included in Pfam (i.e. a novel Pfam domain).
Table 1.
Top ten largest clusters of human regions not covered by Pfam
| Cluster number | Number of regions in the cluster | Region length in amino acids (mean) | Phmmer UniProtKB matches (mean) | Number of phmmer matches with overlaps to Pfam-A families (mean) | Likely annotation |
|---|---|---|---|---|---|
| 1 | 395 | 138 | 27 337 | 4023 | Olfactory receptors |
| 2 | 154 | 121 | 58 170 | 57 777 | Zinc fingers |
| 3 | 104 | 134 | 20 299 | 19 748 | Zinc fingers |
| 4 | 85 | 151 | 17 578 | 14 380 | Collagen repeat |
| 5 | 76 | 127 | 3836 | 3033 | YWTD motifs |
| 6 | 62 | 130 | 2190 | 1958 | Leucine-rich repeats |
| 7 | 56 | 123 | 9662 | 8958 | EGF domains |
| 8 | 46 | 158 | 23 652 | 23 285 | Zinc fingers |
| 9 | 41 | 203 | 701 | 297 | PRY domain |
| 10 | 40 | 178 | 677 | 2 | Cadherin cytoplasmic domains |
Mean number of UniProtKB matches is based on running each region in the cluster against UniProtKB with phmmer. The number of matches with E-value <10−3 is collected, and the average is taken over all regions in a cluster. Overlaps with existing Pfam-A families are calculated based on sequences that match simultaneously a cluster member (according to alignment co-ordinates in phmmer output) and a Pfam-A family (according to alignment co-ordinates in Pfam 27.0). ‘Likely annotation’ is assigned based on analysis of overlapping Pfam clans (when a family is not in a clan, it is considered as being in a clan by itself) and on manual inspection of region annotation in UniProtKB, InterPro (18) and Pfam.
We additionally looked at the top 20 clusters with the largest number of average matches in UniProtKB (excluding those already found in Table 1). These included 12 clusters made up of putative zinc finger domains as well as outliers of other existing families such as the SET family (PF00856), the short-chain dehydrogenase C-terminal domain (PF13561) and an ABC transporter family (PF12848). Examples of putative domain extensions were also present, including putative C-terminal extensions of the Crotonase family (PF00378), a putative N-terminal extension of the cytochrome b N-terminal domain (PF13631), a putative C-terminal extension of the ATP-binding cassette of ABC transporters (PF00005), a putative C-terminal extension of the NADH-Ubiquinone/plastoquinone family (PF00361) and a putative N-terminal extension of the histone family (PF00125).
Finally, it is interesting to note that clusters with a high mean number of phmmer matches in UniProtKB (≥1000) had a high percentage of matches that fell into existing Pfam families (46%). The corresponding figure for clusters with a mean number of matches in UniProtKB lower than 100 was only 4%.
Coverage of the human proteome by structure-based protein family databases
We looked at coverage of our set of human UniProtKB/Swiss-Prot proteins by two structure-based protein family databases: SUPERFAMILY (19) and Gene3D (20) (Methods). These databases build their families starting from experimentally determined structural domains. SUPERFAMILY is based on the SCOP classification (21), and Gene3D is based on the CATH classification (22). SUPERFAMILY had a human sequence coverage of 72% and residue coverage of 41%, while Gene3D covered 69% of human sequences and 35% of human residues. This can be compared with 90% sequence and 45% residue coverage achieved by Pfam-A families. Interestingly, both SUPERFAMILY and Gene3D matched part of the 38% uncovered human residues discussed above. SUPERFAMILY covered 18% of the 38% or an additional 7% of the entire proteome, while Gene3D covered 15% of the 38% or an additional 6% of the entire proteome.
Compositionally biased regions were over-represented in uncovered regions
Next, we looked at the amino acid composition of the portion of the human proteome not covered by Pfam. In Table 2, we compared the percentage of residues predicted to belong to compositionally biased regions in the portion of the human proteome covered by Pfam-A and Pfam-B families and in the portion uncovered by Pfam (excluding from the latter signal peptides and regions shorter than 50 residues). We found that residues predicted to be in low complexity, intrinsically disordered and coiled-coil regions were under-represented in Pfam-A families (note that there is significant overlap between these three categories). Predicted disorder, in particular, was about 4-fold less frequent in Pfam-A families than in the other two categories. We also noted that uncovered human region clusters with a small mean number of matches in UniProtKB (<100) had the highest fraction of disordered residues with respect to all other clusters (43% as compared with, for example, 12% in clusters with ≥1000 matches). In contrast, residues predicted to be part of helical transmembrane regions were 2- to 3-fold more common in Pfam-A families than in the other two categories, while signal peptides were almost totally absent from Pfam-A regions (this was as expected, as we are systematically excluding signal peptides from Pfam families).
Table 2.
Percentage of residues predicted to be compositionally biased in Pfam-A families, Pfam-B families and in regions that are not Pfam-A, not Pfam-B, not predicted to be signal peptides and of at least 50 consecutive amino acids in length
| Pfam-A | Pfam-B | Not (Pfam-A, Pfam-B, signal peptide), ≥50aa | |
|---|---|---|---|
| Coiled-coil | 2.1 | 4.9 | 3.8 |
| Disorder | 9.3 | 42.0 | 38.5 |
| Low complexity | 5.1 | 13.8 | 13.2 |
| Signal peptide | 0.2 | 1.2 | 0 |
| Transmembrane | 6.2 | 1.9 | 2.5 |
Consensus long disordered regions were most over-represented in regions not covered by Pfam
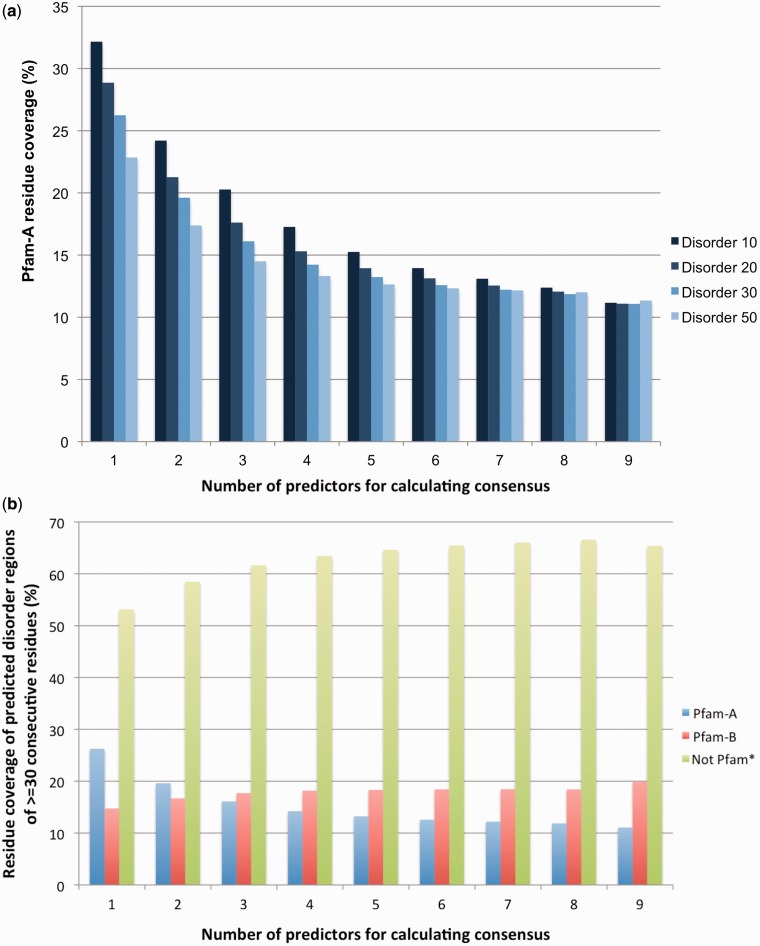
Intrinsically disordered regions in proteins can be described in terms of specific sequence, structural and functional properties that distinguish them from structured domains (23). Intrinsic disorder can take different forms and can overlap with different types of compositional bias. Low complexity regions, coiled–coiled regions, flexible loops and long unstructured regions have all been described, by at least some authors, as disordered. As different prediction methods address different aspects of disorder, it is useful to take into account results from more than one predictor. Here, we used prediction data contained in the D2P2 database (15), which stores predictions from six different methods on 1765 complete proteomes (some methods are used in more than one mode, giving a total of nine sets of predictions for each protein sequence). We were able to successfully map D2P2 human proteome collection to 90% of our set of UniProtKB/Swiss-Prot human proteins (see Methods). Of those sequences successfully mapped to the D2P2 database, 66% of amino acids were predicted by at least one method to be disordered, while 5% were predicted by all methods (consensus set) to be disordered. Pfam-A covers 35% of amino acids predicted by at least one method and 11% of those in the consensus set, respectively. In Figure 3a, we looked at predicted consecutive disordered regions of increasing length (from 10 to 50 residues) and reported the Pfam-A residue coverage of such regions. Predicted disordered regions were defined by increasing consensus in D2P2 (from 1 prediction to 9 predictions). Pfam-A coverage decreased with increasing length of the disordered region and with increasing consensus between the different predictors. In Figure 3b, we focussed on predicted consecutive disordered regions of 30 or more residues and compared Pfam-A, Pfam-B and non-Pfam regions. While 26% (one predictor) and 11% (all nine predictors) of regions were found in Pfam-A (Figure 3b, blue), for Pfam-B families and non-Pfam regions, we observed the opposite trend (Figure 3b, red and green, respectively).
Figure 3.
(a) Pfam-A residue coverage of consensus predicted human disordered regions as a function of region length and number of predictors considered for consensus. Predicted disordered regions of different lengths are considered. Disorder X stands for regions of at least X consecutive disordered amino acids. Only regions of length X that are predicted in full by N predictors are considered (N= 1 , .. , 9; x-axis). For example, 15% of the residues found in regions of 10 consecutive predicted disordered residues by at least five different methods are found in Pfam-A families. (b) Comparison of human residue coverage of predicted disordered regions of length 30 for Pfam and not Pfam. ‘not Pfam*’ represents all regions of length ≥50 residues that are not in Pfam-A, not in Pfam-B and not predicted to be signal peptides.
Discussion
Improving existing Pfam-A families can lead to significant gains in residue coverage of the human proteome
A total of 5494 Pfam-A families have at least one match in the UniProtKB/Swiss-Prot collection of human proteins; 1937 of these (or 35%) are found in 395 Pfam clans. Overall, 90% of human sequences have at least one Pfam-A match. We are thus very close to having at least one Pfam-A conserved region for every human protein. However, providing high coverage at the amino acid level, now at ∼45%, is likely to be a challenge. About 10% of additional coverage could be obtained by using the 9418 Pfam-B families that contain human protein regions as a starting point to build new Pfam-A families. Even if these families were successfully built, 38% of the residues in the human proteome would remain uncovered by Pfam-A (after additionally excluding signal peptides and regions shorter than 50 amino acids). This ‘uncovered’ portion of the human proteome is split across almost 25 000 regions that we have grouped into >15 000 homologous clusters. Our analysis suggests that uncovered human regions with a high number of homologues in UniProtKB most often fall into one of two categories: (i) divergent members of existing Pfam families or (ii) N- or C-terminal extensions of existing Pfam families. In the first case, the uncovered regions could be recovered by either improving the alignment used to generate the family profile HMM, or by adding a new family to an existing clan. Alternatively, when dealing with short motifs or repeat families, integrating these regions may become possible through future HMMER algorithmic developments. In the second case, covering regions such as the N-terminal portions of the olfactory receptor family PF13853 discussed in the Results section will require extending the boundaries of existing Pfam-A families so that these represent full functional/structural domains. Almost 2/3 of the clusters that we identified that account for 34% of all uncovered regions, however, appear to have only a small number of homologous regions in UniProtKB (less than 100 using phmmer; note, however, that using jackhmmer iterative searches may return more matches). Seventy-four percent of these clusters have members that align exclusively to UniProtKB regions not covered by Pfam-A and hence represent potential new Pfam families. The fact that members of these clusters have a high percentage of disordered residues (43%), however, raises the question as to whether homologous inference will be accurate for these regions. This makes their classification all the more challenging, as we discuss in the next section. Finally, comparison with coverage provided by structure-based protein family databases indicates that at least some new human Pfam families could be built from and some existing Pfam families improved with the help of structural information.
Most uncovered regions rich in intrinsically disordered amino acids
A significant fraction of residues that are not yet covered by Pfam-A are predicted by different algorithms to be located in disordered regions (Figure 3b). We have also seen that the longer the disordered regions, the more under-represented they are in Pfam-A (Figure 3a). It should be noted that under-representation of disorder in Pfam-A is not a phenomenon specific to the human proteome but applies to the whole of UniProtKB. This helps to explain the huge gap between residue coverage in E. coli and residue coverage in human and S. cerevisiae (Figure 1), as disorder is known to be much more common in Eukaryotes than it is in Bacteria (24) [see also the D2P2 (15) and MobiDB (25) databases]. Until a few years ago, intrinsically disordered regions had attracted little attention from experimental biologists and bioinformaticians alike. More recently, intrinsic disorder has taken centre stage (26), with the establishment of databases that collect experimental evidence for disorder (27, 28) and the development of dozens of computational methods to predict it (29). Disordered regions are known to be involved primarily in cell signalling and regulation (30) and have been linked to disease (31, 32).
A possible explanation for the small number of disordered regions found in Pfam-A is that disordered regions seem, on average, to be less conserved than their structured counterparts. There are cases of Pfam-A families that include protein regions experimentally shown to be fully disordered. For example, family PF05456 (eIF_4EBP) includes the human eukaryotic translation initiation factor 4E-binding protein 1 (Q13541), which has been shown by nuclear magnetic resonance experiments to be fully disordered (33). However, while some intrinsically disordered regions appear to be conserved in sequence (34, 35), it is not yet clear how true this is in general, nor how good alignment methods are at detecting homology in these compositionally biased regions. The latter observation is especially relevant in the case of long disordered regions (34). Forslund and Sonnhammer’s benchmarking of the efficacy of BLAST filters for homology recognition in low complexity regions (36), although encouraging, was based on regions predicted by the program SEG (37), which represent only one particular flavour of disorder. In conclusion, it is possible that many of the uncovered human regions that are significantly enriched in disorder, such as the ones with few homologues in UniProtKB, will turn out to be too difficult to incorporate into the Pfam classification.
Future Pfam strategies
We plan to adopt a two-pronged strategy for increasing coverage of the human proteome. Firstly, as we believe the examples discussed here have convincingly shown, there is ample space and purpose for improving existing human-member families. This should provide us with a better coverage of their homologous UniProtKB regions and improve their correspondence to full functional/structural domains. Secondly, when building new Pfam-A families, we will prioritize regions for which functional and/or structural information is available, thus increasing the chance of correctly assigning family boundaries. Manual inspection of alignments should also ensure that intrinsically disordered regions are put into families only when they exhibit clear amino acid-conservation patterns.
Conclusions
We analyse regions of the human proteome currently falling outside of the Pfam classification of protein families. We see that ∼38% of amino acids in the human proteome are not in Pfam-A or Pfam-B families and are found in regions ≥50 amino acids. Analysis of these regions shows that for the purpose of increasing coverage of the human proteome, improving existing families may be as important as building new ones. Also, uncovered regions that do not appear to be related to existing families exhibit on average a significant percentage of residues predicted to be in intrinsically disordered regions. Given the difficulty in correctly aligning compositionally biased disordered regions, incorporating them into Pfam is likely to be challenging. Based on these findings, we propose a Pfam strategy for increasing coverage of the human proteome that aims at improving existing families and building new ones primarily from regions that have been experimentally functionally and/or structurally characterized.
Acknowledgements
We are grateful for the infrastructure support provided by the Systems, Web and Database administration teams at Wellcome Trust Sanger Institute (WTSI). M.P. and J.M. would like to thank Julian Gough and Matt Oates (both University of Bristol) for help with using the D2P2 database and Craig McAnulla (EBI) for help in running the latest version of InterProScan. M.P. would like to thank Lars Barquist (WTSI) for useful discussions.
Funding
Wellcome Trust (WT077044/Z/05/Z). BBSRC Bioinformatics and Biological Resources Fund (BB/F010435/1). Howard Hughes Medical Institute (to R.D.F.). Funding for open access charge: Wellcome Trust (WT077044/Z/05/Z).
Conflict of interest. None declared.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Dunham I, Kundaje A, Aldred SF, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dimmer EC, Huntley RP, Alam-Faruque Y, et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2012;40:D565–D570. doi: 10.1093/nar/gkr1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heger A, Wilton CA, Sivakumar A, et al. ADDA: a domain database with global coverage of the protein universe. Nucleic Acids Res. 2005;33:D188–D191. doi: 10.1093/nar/gki096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finn RD, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machida K, Mayer BJ. The SH2 domain: versatile signaling module and pharmaceutical target. Biochim. Biophys. Acta. 2005;1747:1–25. doi: 10.1016/j.bbapap.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Mayer BJ. SH3 domains: complexity in moderation. J. Cell. Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 9.Zarrinpar A, Bhattacharyya RP, Lim WA. The structure and function of proline recognition domains. Sci. STKE. 2003;2003:RE8. doi: 10.1126/stke.2003.179.re8. [DOI] [PubMed] [Google Scholar]
- 10.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 12.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput. Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]
- 13.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Dosztanyi Z, Csizmok V, Tompa P, et al. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 15.Oates ME, Romero P, Ishida T, et al. D2P2: database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–DD516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keseler IM, Collado-Vides J, Santos-Zavaleta A, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherry JM, Hong EL, Amundsen C, et al. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunter S, Jones P, Mitchell A, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gough J, Karplus K, Hughey R, et al. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 20.Lees J, Yeats C, Perkins J, et al. Gene3D: a domain-based resource for comparative genomics, functional annotation and protein network analysis. Nucleic Acids Res. 2012;40:D465–D471. doi: 10.1093/nar/gkr1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreeva A, Howorth D, Chandonia JM, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orengo CA, Martin AM, Hutchinson G, et al. Classifying a protein in the CATH database of domain structures. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1155–1167. doi: 10.1107/s0907444998007501. [DOI] [PubMed] [Google Scholar]
- 23.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pancsa R, Tompa P. Structural disorder in eukaryotes. PLoS One. 2012;7:e34687. doi: 10.1371/journal.pone.0034687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Domenico T, Walsh I, Martin AJ, et al. MobiDB: a comprehensive database of intrinsic protein disorder annotations. Bioinformatics. 2012;28:2080–2081. doi: 10.1093/bioinformatics/bts327. [DOI] [PubMed] [Google Scholar]
- 26.Dunker AK, Oldfield CJ, Meng J, et al. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9 (Suppl. 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sickmeier M, Hamilton JA, LeGall T, et al. DisProt: the database of disordered proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuchi S, Sakamoto S, Nobe Y, et al. IDEAL: intrinsically disordered proteins with extensive annotations and literature. Nucleic Acids Res. 2012;40:D507–D511. doi: 10.1093/nar/gkr884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He B, Wang K, Liu Y, et al. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 30.Tantos A, Han KH, Tompa P. Intrinsic disorder in cell signaling and gene transcription. Mol. Cell Endocrinol. 2012;348:457–465. doi: 10.1016/j.mce.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 31.Babu MM, van der Lee R, de Groot NS, et al. Intrinsically disordered proteins: regulation and disease. Curr. Opin. Struct. Biol. 2011;21:432–440. doi: 10.1016/j.sbi.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 32.Midic U, Oldfield CJ, Dunker AK, et al. Unfoldomics of human genetic diseases: illustrative examples of ordered and intrinsically disordered members of the human diseasome. Protein Pept. Lett. 2009;16:1533–1547. doi: 10.2174/092986609789839377. [DOI] [PubMed] [Google Scholar]
- 33.Fletcher CM, Wagner G. The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci. 1998;7:1639–1642. doi: 10.1002/pro.5560070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen JW, Romero P, Uversky VN, et al. Conservation of intrinsic disorder in protein domains and families: I. A database of conserved predicted disordered regions. J. Proteome. Res. 2006;5:879–887. doi: 10.1021/pr060048x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bellay J, Han S, Michaut M, et al. Bringing order to protein disorder through comparative genomics and genetic interactions. Genome Biol. 2011;12:R14. doi: 10.1186/gb-2011-12-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forslund K, Sonnhammer EL. Benchmarking homology detection procedures with low complexity filters. Bioinformatics. 2009;25:2500–2505. doi: 10.1093/bioinformatics/btp446. [DOI] [PubMed] [Google Scholar]
- 37.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]