Abstract
When annotating protein sequences with the footprints of evolutionarily conserved domains, conservative score or E-value thresholds need to be applied for RPS-BLAST hits, to avoid many false positives. We notice that manual inspection and classification of hits gathered at a higher threshold can add a significant amount of valuable domain annotation. We report an automated algorithm that ‘rescues’ valuable borderline-scoring domain hits that are well-supported by domain architecture (DA, the sequential order of conserved domains in a protein query), including tandem repeats of domain hits reported at a more conservative threshold. This algorithm is now available as a selectable option on the public conserved domain search (CD-Search) pages. We also report on the possibility to ‘suppress’ domain hits close to the threshold based on a lack of well-supported DA and to implement this conservatively as an option in live conserved domain searches and for pre-computed results. Improving domain annotation consistency will in turn reduce the fraction of NR sequences with incomplete DAs.
Introduction
The Conserved Domain Database (CDD) (1) consists of a collection of well-annotated multiple sequence alignment models for ancient domains and full-length proteins. These models are available as position-specific score matrices (PSSMs) for identifying conserved domains in protein sequences via Reverse Position-Specific (RPS)-BLAST. CDD is a redundant collection, it includes models imported from SMART (2), Pfam (3), COG (4), NCBI Protein Clusters (5) and TIGRFAMS (6), as well as NCBI-curated fine-grained hierarchical classifications for selected domain families based on phylogenetic analysis. Domain models that have overlapping annotation on the same protein sequences are clustered into CDD superfamilies (7). CDD provides pre-computed domain and site annotation for the majority of protein sequences tracked by NCBI’s Entrez system. Two CDD search services are available: CD-Search (8), for protein and nucleotide queries and Batch CD-Search (9), for multiple protein queries. The default E-value threshold for the pre-computed annotation and for these search services is 0.01. NCBI’s Conserved Domain Architecture Retrieval Tool (CDART) (10) carries out similarity searches of Entrez Protein based on domain architecture (DA). For a protein query, it returns the footprints of the highest scoring CDD superfamilies on that protein sequence and a list of proteins with similar DAs, grouped according to DA. CDART output for each DA includes its taxonomy span and the total number of NR sequences having that DA.
Methods
Estimating the frequency of domain hits to ‘rescue’
It has been noted earlier that profile-based annotation of domain footprints can benefit from considering domain co-occurrence (11). To determine whether developing an algorithm to ‘rescue’ domain hits above the default reporting threshold of E-value 0.01 would uncover a significant number of additional annotations, we estimated the frequency of domain hits we would ‘rescue’ by manually inspecting randomly picked sets of sequences. We chose SwissProt (12) as represented in NCBI’s Entrez/protein database (542 902 sequences) and a representative human proteome (19 856 sequences, with ‘NP’ and ‘XP’ accession prefixes) as test sets (5 February 2014). The human proteome comprised essentially one representative protein sequence for each currently known human gene. To generate the set, we parsed the annotation files for the human reference assembly GRCh38 for GeneID and protein accessions and applied selection criteria to pick one per gene based on the longest annotated CDS per GeneID. We obtained three random subsets of 4000 sequences from SwissProt and two random subsets of 2000 sequences each from the representative human proteome and compared the domain hits reported at E-value thresholds of 1.0 vs. the default reporting threshold at 0.01 for these smaller subsets of proteins. An in-house version of CDART was generated that contains pre-computed sets of RPS-BLAST results for all sequences in NCBI’s NR database reported at an E-value threshold of 1.0, above the default reporting threshold of 0.01. We asked how often new or additional domain hits were encountered and how often this happens when other non-overlapping domain footprints are present. We manually inspected subsets of the protein sequences that had found new hits to determine whether these hit(s) appeared meaningful and should be ‘rescued’. In making that decision, we considered the frequency and taxonomy of the new DA, completeness of the ‘new’ domain hit and overlap between the ‘new’ domain hit and other well-supported domain annotations.
Consequently, we generated a set of protein sequences having valuable domain hits to ‘rescue’ and used it to later refine the following ‘rescue’ algorithm based on well-supported DA or tandem repeats.
‘Rescue algorithm’
Report the search results with the default E-value threshold (currently 0.01) and record the domain hits—defined as the ‘A’ domain hits.
Report the search results with the increased E-value threshold (such as 1.0). There may now be additional domain hits—defined as the ‘B’ domain hits.
Record the frequency of the DA formed by the ‘A’ domain hits.
Record the frequency of all alternative DAs that contain all of the ‘A’ domains and all combinations of the additional ‘B’ domain hits but no additional domain hits that are not included in the ‘A’ and ‘B’ sets.
Rank the alternative DAs by frequency. If the most frequent alternative covers at least 20 sequences from NCBI’s NR database or is more common than the initial ‘A’ DA, report it instead of the ‘A’ architecture, which means that ‘rescued’ domains that contribute to this most frequent DA are reported as well.
As CDART only reports/considers a single superfamily footprint for two or more consecutive domain hits to models from the same superfamily, irrespective of the repeat number, we added the following: additionally, if any of the additional domain hits that has not been ‘rescued’ at this point belongs to the same CDART superfamily as an adjacent hit that is being reported (i.e., a tandem repeat), that domain hit is being ‘rescued’ and reported as well.
Results and conclusions
Improving domain annotation consistency to increase the fraction of sequences in NR with more complete DAs
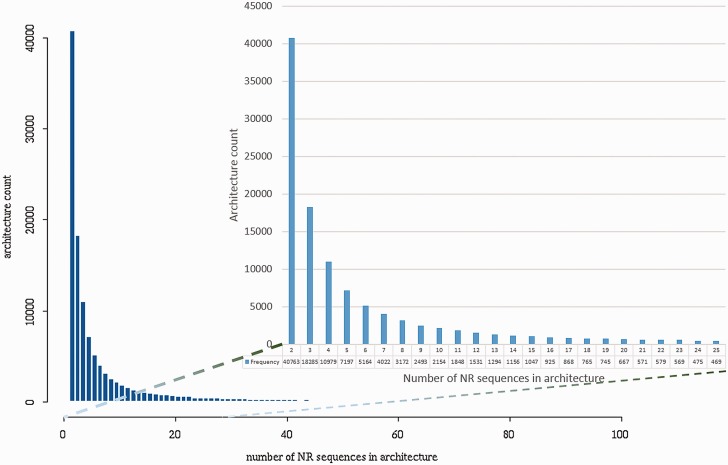
Incomplete architectures may contribute considerably to the large overall number of DAs in CDART. As shown in Figure 1, a large fraction of DAs each cover only a few sequences from NCBI’s NR protein set. On-going curation to improve representation of some domain families and other efforts such as reported here to improve domain annotation consistency will reduce the fraction of sequences with incomplete DAs and hence the number of rare or unusual DAs in CDART.
Figure 1.
DAs in CDART and their corresponding number of sequences in the NR database. Determined 6 June 2014.
Manual inspection adds a non-trivial amount of valuable annotation using CDD at a higher threshold
We manually screened random subsets of protein sequences having one or more additional domain hits at RPS-BLAST E-value threshold of 1.0 which were not present at the reporting E-value threshold of 0.01 and determined whether these additional hit(s) were valid and should be ‘rescued’ (Table 1). For example, in the SwissProt random sample 2, 987 of the 4000 unique protein sequences had new hits (24.7% of the total protein sequences). Of these 987 protein sequences, about 19.5% had valuable hits to ‘rescue’, which can be extrapolated to about 4.8% of the original 4000 random sample. Information such as the frequency of the alternative DA, the taxonomical span of the alternative DA, the completeness of the additional domain hit and its degree of overlap with other well-supported domain hits factored into the decision to ‘rescue’ these additional hits. We determined that approximately 5% and 19.5% of protein sequences have valid domain hit(s) to ‘rescue’ for the SwissProt and human proteome test sets, respectively (Table 1, average and standard deviation: 4.97% ± 0.13% and 19.66% ± 5.18%). Even at about 4.81% (the lower percentage determined from random samples having valid hits), this translates into a non-trivial number of protein sequences in NR receiving valuable annotation that is currently not seen in pre-computed results or in CD searches run at the default E-value threshold of 0.01. A significant portion of the domains to ‘rescue’ are tandem repeats; 1.23% ± 0.19% and 7.36% ± 0.18% of protein sequences have valid tandem repeats to ‘rescue’ for the SwissProt and human proteome test sets, respectively.
Table 1.
Manual inspection adds a non-trivial amount of valuable annotation using CDD at a higher threshold
| CDD V3.11 45 746 PSSMs. | Human sample A: 2000 protein sequences | Human sample B: 2000 protein sequences | SwissProt sample 1: 4000 protein sequences | SwissProt sample 2: 4000 protein sequences | SwissProt sample 3: 4000 protein sequences |
|---|---|---|---|---|---|
| Percentage having additional hits at E-value 1 | 48.00 | 48.25 | 24.00 | 24.68 | 24.58 |
| Percentage having additional hits at E-value 1 to ‘rescue’ | 16.00 | 23.32 | 5.06 | 4.81 | 5.02 |
| Percentage having additional tandem repeat hits at E-value 1 to ‘rescue’ | 7.48 | 7.24 | 1.27 | 1.40 | 1.02 |
An algorithm that considers DA and tandem repeats adds a significant amount of valuable annotation using CDD at a higher threshold
An automated procedure (detailed in the Methods section) was developed, a simple classifier which considers DA and tandem repeats in making the determination as to whether to ‘rescue’ a borderline hit; 11.29% of protein sequences in the representative human proteome (2241 protein sequences out of 19 856) and 5.58% of the SwissProt protein sequences (30 267 protein sequences out of 542 902) had domain(s) ‘rescued’ by the algorithm. In the manual screening (Table 1), we estimated 19.66% ± 5.18% and 4.97% ± 0.13% and (averages and standard deviations) for the human proteome and SwissProt test sets, respectively. However, in the manual screening, we considered additional discriminators, such as taxonomy of the DA in CDART, and different thresholds of NR sequences and cannot exclude unconscious bias based on the biological knowledge of the curator. These additional discriminators were not considered in the automated procedure. In addition, the algorithm was run on a later CDD version/release CDD V3.12 46 675 PSSMs with improved domain models. This may explain the difference in the percent of rescued domains between our manual screening and automated procedure. From this study, it may not be necessary to include additional and more computationally intensive discriminators in the algorithm, such as taxonomic distribution.
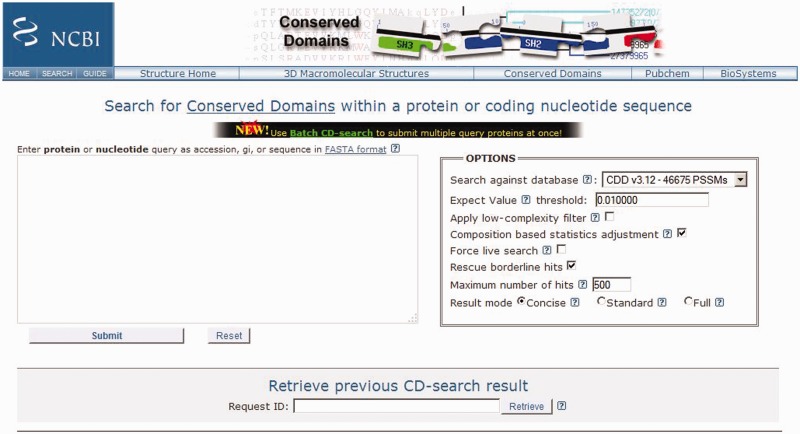
The automated algorithm was implemented in the latest public CD-Search version (released 3 October 2014), as a selectable option ‘rescue borderline hits’ for live searches (Figure 2).
Figure 2.
The public CD-Search (release 3 October 2014) now supports the ‘rescue’ of borderline-scoring domain hits based on well-supported DA, for live searches.
Example of a domain ‘rescued’ by the algorithm and of an incomplete DA
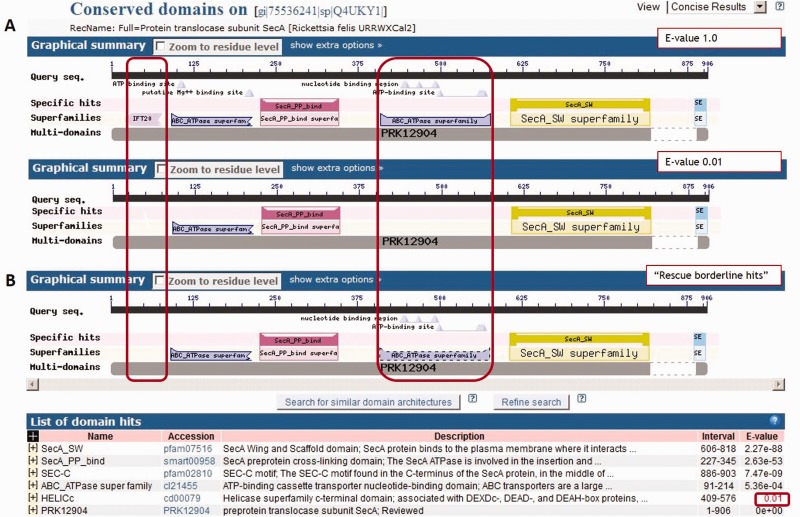
Figure 3 shows an example of the algorithm as applied to Rickettsia felis protein translocase subunit SecA (GI:75536241). Figure 3A shows live CD-Search results. There were four A domain hits: A1 = ABC_ATPase, A2 = SEcA_PP_bind, A3 = SecA_SW, A4 = SecA_C and two B domain hits (B1 = IFT20 and B2 = ABC_ATPase). Searching at the reporting threshold gives [A1]-[A2]-[A3]-[A4], and searching at the raised E-value 1.0 gives -[B1]-[A1]-[A2]-[B2]-[A3]-[A4]-. The frequency of the DA from CDART at the reporting threshold E-value is shown in Table 2. Figure 3B shows the results of a live CD-Search result selecting the new ‘rescue borderline hits’ option. Only one of the two new hits detected at the raised E-value is ‘rescued’, B2 = ABC_ATPase, it is indicated by a dotted line and its E-value is highlighted in red. Based on DA, the ABC_ATPase superfamily domain hit (cl21455, hit detected with superfamily member cd00079 HELICc) is ‘rescued’ by the algorithm, the intraflagellar transport complex B subunit 20 (IFT20) domain hit (cl20817) is not.
Figure 3.
Rickettsia felis SecA (GI:75536241): an example domain hit ‘rescued’ by the algorithm (computed 4 November 2014).
Table 2.
Rickettsia felis SecA (GI:75536241) example: DA frequency in CDART at reporting threshold
| DA frequency at E-value 0.01 (NR sequences) | |
|---|---|
| -[A1]-[A2]-[A3]-[A4] | 1542 |
| Alternative DAs | |
| -[A1]-[A2]-[B2]-[A3]-[A4] | 2554 |
| -[B1]-[A1]-[A2]-[B2]-[A3]-[A4] | 0 |
There are two common DAs in CDART, one with and one without the second ABC_ATPase SF domain (Table 2, 2554 vs. 1542 NR sequences, respectively, determined 4 November 2014). The DA missing the second ATPase domain is most likely incomplete, as the SecA ATPase/DEAD motor composed of two ATPase domains, which function together to bind and hydrolyze ATP. For about 80% of the sequences in NR having the DA lacking the second ATPase domain (as of 16 October 2014), a CD-Search with the new ‘Rescue borderline hit’ option ‘rescued’ the second ATPase domain. It may be that with improved domain representation and detection, and with improved annotation consistency, these two architectures will resolve to a single DA with two ATPase domains.
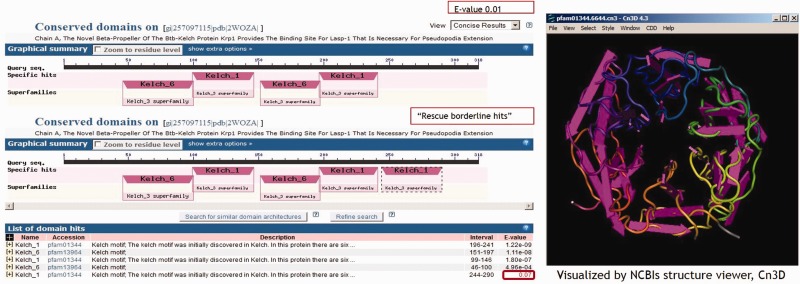
Example of tandem repeats lifted by the algorithm
The algorithm also ‘rescues’ all additional hits detected at E-value 1 and not at E-value 0.01 that belong to the same CDART superfamily as an adjacent hit that is being reported at E-value 0.01, i.e. tandem repeats. An example (Figure 4) is the beta-Propeller of protein Krp1, which contains six Kelch repeats. Various pfam models detect four of the six Kelch repeats at the reporting threshold of E-value 0.01 and the fifth Kelch repeat at the raised threshold of E-value 1. The sixth repeat formed from the most C-terminus of the protein and from the N-terminus was undetected.
Figure 4.
The algorithm ‘rescues’ valuable borderline-scoring domain hits if they are tandem repeats of domain hit(s) already reported.
Manual inspection removes a considerable amount of incorrect annotation using CDD at a lower threshold
We were also interested in whether some annotation with borderline hits close to the reporting threshold should be ‘suppressed’. To investigate this, we manually screened sample protein sequences for domain(s) present at the default E-value threshold of 0.01 but lost at an E-value threshold of 0.001 and determined if those domain hits should be ‘suppressed’ (Table 3). For example, of the 2000 random sample D, 260 unique protein sequences lost hits at E-value 0.001 (13% of the total protein sequences). Of these 13%, 2.11% had apparent false-positive hits to ’suppress’, which translates to 0.27% of the starting random sample. We considered the frequency of the alternative DA, the taxonomy of the alternative DA, the completeness of the lost domain hit and its degree of overlap with other well-supported domain hits. Approximately 0.5% and 1.2% of sequences have domains that should be ‘suppressed’ in the human proteome and Swissprot test sets, respectively (Table 3, average and standard deviation: 0.47% ± 0.27% and 1.17% ± 0.45). Even at the lowest percent detected (0.27%), this translates to a non-trivial number of sequences in NR receiving incorrect annotation at E-value 0.01 that ideally should be ‘suppressed’.
Table 3.
Manual inspection removes a significant amount of incorrect annotation using CDD at a lower threshold
| Human sample C: 2000 protein sequences | Human sample D: 2000 protein sequences | SwissProt sample 4: 4000 protein sequences | SwissProt sample 5: 4000 protein sequences | SwissProt sample 6: 4000 protein sequences | |
|---|---|---|---|---|---|
| Percentage having lost hit(s) at E-value 0.001 | 13.00 | 13.00 | 5.38 | 4.75 | 5.18 |
| Percentage having hits to ‘suppress’ | 0.66 | 0.27 | 1.39 | 0.66 | 1.47 |
Example of a domain hit that manual inspection classifies as incorrect
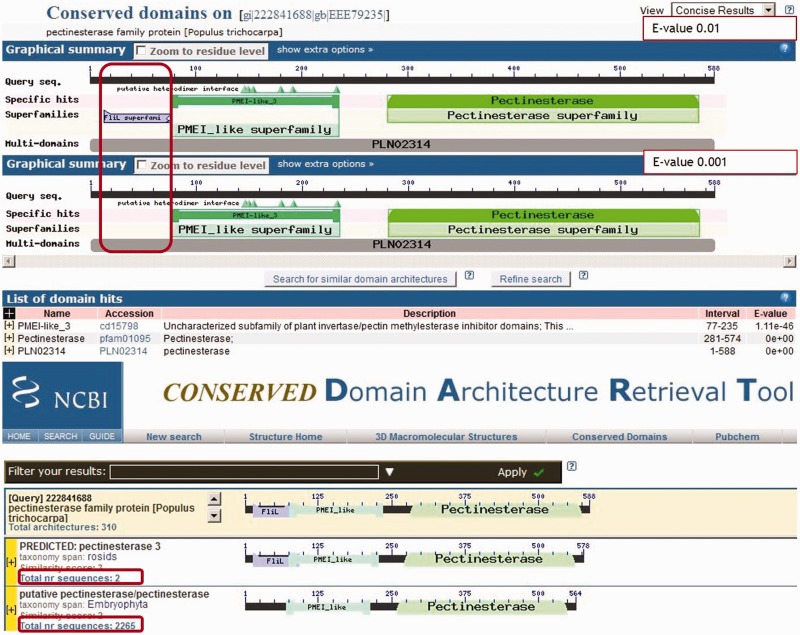
Figure 5 is an example of a borderline hit close to the reporting threshold that should be ‘suppressed’: the FliL (cl00681) hit on Plant (Poplar) Pectinesterase family protein (GI: 222841688). The FliL hit is fragmentary, overlaps the much stronger hit to the plant invertase/pectin methylesterase inhibitor (PME1) domain and being bacterial, it looks out of place in a plant protein. Only two sequences in NR (as of 9 January 2015) have a DA having FliL in combination with both PME1 and Pectinesterase domains.
Figure 5.
Pectinesterase family protein (GI: 222841688): example of an incorrect domain hit that that should be ‘suppressed’.
In summary
Manual inspection (i) reveals a non-trivial amount of valuable annotation using CD-Search at a higher E-value threshold and (ii) also reveals a smaller, but non-trivial amount of incorrect annotation that could be avoided using CD-Search at a lower threshold. The most recent version of CD-Search (released 3 October 2014) provides the option to ‘rescue’ borderline-scoring domain hits based on well-supported DAs and tandem repeats. Currently, this option is available for live searches. We plan to extend the post-processing of CD-Search results to also allow ‘suppression’ of some domains close to the default E-value threshold based on well-supported DA and finally to implement a conservative post-processing strategy for both pre-computed results and live searches.
Acknowledgements
We thank the Conserved Domain Curators past and present for compiling the domain annotations analyzed in this work, Farideh Chitsaz, Marc Gwadz, Fu Lu, James Song, Narmada Thanki, Roxanne Yamashita, Chanjuan Zheng and the CDD alumni Anastasia Nikolskaya, Raja Mazumder, Natalie Fedorova, Aviva Jacobs, B. Sridhar Rao, Sona Vasudevan, Luning Hao, Jodie Yin, Dmitri Krylov, Asba Tasneem, Zhaoxi Ke, Mikhail Mullokandov, Marina Omelchenko, John Jackson, John Anderson, Cynthia Robertson and Carol DeWeese-Scott.
Funding
Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Funding for open access charge: Intramural Research Program of the National Library of Medicine at the National Institutes of Health/DHHS.
Conflict of interest: None declared.
References
- 1.Marchler-Bauer A., Zheng C., Chitsaz F., et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res., 41(Database Issue), D348–D352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic I., Doerks T., Bork P. (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res., 43(Database Issue), D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finn R.D., Bateman A., Clements J., et al. (2014) Pfam: the protein families database. Nucleic Acids Res., 42(Database Issue), D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatusov R.L., Natale D.A., Garkavtsev I.V., et al. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klimke W., Agarwala R., Badretdin A., et al. (2009) The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res., 37(Database issue), D216–D223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haft D.H., Selengut J.D., Richter P.A., et al. (2013) TIGRFAMS and genome properties in 2013. Nucleic Acids Res., 41(Database Issue), D387–D395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchler-Bauer A., Anderson J.B., Chitsaz F., et al. (2009) CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res., 37(Database Issue), D205–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchler-Bauer A., Bryant S.H. (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res., 32, 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchler-Bauer A., Lu S., Anderson J.B., et al. (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res., 39(Database Issue), D225–D229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geer L.Y., Domrachev M., Lipman D.J., et al. (2003) CDART: protein homology by domain architecture. Genome Res., 12, 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coin L., Bateman A., Durbin R. (2003) Enhanced protein domain discovery by using language modeling techniques from speech recognition. Proc. Natl Acad. Sci. USA, 100, 4516–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res., 43(Database Issue), D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]