ABSTRACT
African swine fever (ASF) is a highly infectious disease caused by the African swine fever virus (ASFV) in swine. It is characterized by the death of cells in infected tissues. However, the molecular mechanism of ASFV-induced cell death in porcine alveolar macrophages (PAMs) remains largely unknown. In this study, transcriptome sequencing of ASFV-infected PAMs found that ASFV activated the JAK2-STAT3 pathway in the early stages and apoptosis in the late stages of infection. Meanwhile, the JAK2-STAT3 pathway was confirmed to be essential for ASFV replication. AG490 and andrographolide (AND) inhibited the JAK2-STAT3 pathway, promoted ASFV-induced apoptosis, and exerted antiviral effects. Additionally, CD2v promoted STAT3 transcription and phosphorylation as well as translocation into the nucleus. CD2v is the main envelope glycoprotein of the ASFV, and further investigations showed that CD2v deletion downregulates the JAK2-STAT3 pathway and promotes apoptosis to inhibit ASFV replication. Furthermore, we discovered that CD2v interacts with CSF2RA, which is a hematopoietic receptor superfamily member in myeloid cells and a key receptor protein that activates receptor-associated JAK and STAT proteins. In this study, CSF2RA small interfering RNA (siRNA) downregulated the JAK2-STAT3 pathway and promoted apoptosis to inhibit ASFV replication. Taken together, ASFV replication requires the JAK2-STAT3 pathway, while CD2v interacts with CSF2RA to regulate the JAK2-STAT3 pathway and inhibit apoptosis to facilitate virus replication. These results provide a theoretical basis for the escape mechanism and pathogenesis of ASFV.
IMPORTANCE African swine fever is a hemorrhagic disease caused by the African swine fever virus (ASFV), which infects pigs of different breeds and ages, with a fatality rate of up to 100%. It is one of the key diseases affecting the global livestock industry. Currently, no commercial vaccines or antiviral drugs are available. Here, we show that ASFV replicates via the JAK2-STAT3 pathway. More specifically, ASFV CD2v interacts with CSF2RA to activate the JAK2-STAT3 pathway and inhibit apoptosis, thereby maintaining the survival of infected cells and promoting viral replication. This study revealed an important implication of the JAK2-STAT3 pathway in ASFV infection and identified a novel mechanism by which CD2v has evolved to interact with CSF2RA and maintain JAK2-STAT3 pathway activation to inhibit apoptosis, thus elucidating new information regarding the signal reprogramming of host cells by ASFV.
KEYWORDS: ASFV, JAK2-STAT3 pathway, CD2v, CSF2RA, apoptosis
INTRODUCTION
African swine fever (ASF) is a highly contagious and hemorrhagic infectious disease caused by the African swine fever virus (ASFV), which infects pigs of different breeds and ages, with a fatality rate of up to 100%. It is one of the most important diseases affecting the global pig industry. Currently, there are no commercially available vaccines or antiviral drugs for ASFV infection (1–3). It is a large, complex double-stranded DNA double-membrane virus (4). The size of the ASFV genome ranges from 170 to 194 kb, the diameter of the virions is approximately 200 nm, and the genome contains 151 to 167 open reading frames (ORFs). ASFV carries at least 160 proteins, of which approximately 50 proteins currently have known functions and play an important role in evading host defense (5–7). CD2v, a glycoprotein in the ASFV envelope, can mediate the hemadsorption (HAD) of red blood cells, induce a protective immune response, participate in immune escape, and enhance virus replication in soft ticks (8–12). Therefore, the interaction between CD2v and the host may regulate the immune escape and pathogenesis of ASFV; however, the target proteins and underlying mechanisms remain unclear.
To better understand the pathogenesis of host regulation by ASFV infection, transcriptomic sequencing results of ASFV-infected porcine alveolar macrophages (PAMs) were analyzed, and it was found that ASFV infection activated the JAK2-STAT3 pathway. The JAK2-STAT3 pathway is a proinflammatory cytokine-induced pathway, also known as the enhanced survival activator (SAFE) pathway (13). It is activated by various cytokines, including interleukins (ILs), colony-stimulating factors, and interferons, and triggers inflammation. The JAK2-STAT3 pathway is involved in cell proliferation, differentiation, apoptosis, and inflammation (14). Studies have shown that the JAK2-STAT3 pathway can upregulate the expression levels of the antiapoptotic proteins BclII and Bcl-xL. Inhibition of the JAK2-STAT3 pathway has been reported to significantly inhibit the replication of the influenza A virus (15, 16), porcine epidemic diarrhea virus (17), and hepatitis A virus (18). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) induces hyperactivation of proinflammatory cytokines through the IL-6-JAK-STAT3 pathway, and inhibition of the JAK2-STAT3 pathway can control SARS-CoV-2-generated cytokine storms (19). Spinal cord injury inhibits inflammation and promotes disease recovery by inhibiting the JAK2-STAT3 pathway (20). Overexpression of the TFPI gene can induce apoptosis of vascular smooth muscle cells by inhibiting the expression of the antiapoptotic protein BclII by inhibiting the JAK2-STAT3 pathway (21). In the glioma cell line U251, Ad-bFGF-small interfering RNA (siRNA) blocks the activation of JAK2 and reduces IL-6 secretion and STAT3 phosphorylation, leading to a reduced expression level of downstream antiapoptotic protein Bcl-xL, ultimately leading to mitochondrial membrane potential disorder and induction of apoptosis (22).
To further explore the mechanism of the JAK2-STAT3 pathway in ASFV infection, the ASFV CD2v protein, which significantly upregulates STAT3 transcription, was screened by a luciferase reporter assay. CD2v also promotes STAT3 phosphorylation in the nucleus, and the CD2v-CSF2RA interaction, initially identified by the yeast two-hybrid assay, was confirmed by a coimmunoprecipitation assay. The CSF2RA protein is a member of the hematopoietic receptor superfamily in bone marrow cells. CSF2RA can activate the signal transduction and activator of receptor-related Janus tyrosine kinase (JAK), transcription (STAT) protein, and mitogen-activated protein kinase (MAPK) signal transduction pathways (23). It has been reported that activation of the GM-CSFRα promoter induces GM-CSFRα production, which protects CLL cells from apoptosis (24). Overexpression of GM-CSFRα protects MM1 cells from dexamethasone-induced apoptosis by phosphorylating STAT3 (25). Hence, the principal focus of this study was to determine how the ASFV CD2v protein regulates virus replication.
RESULTS
The ASFV GZ201801 strain induces host apoptosis.
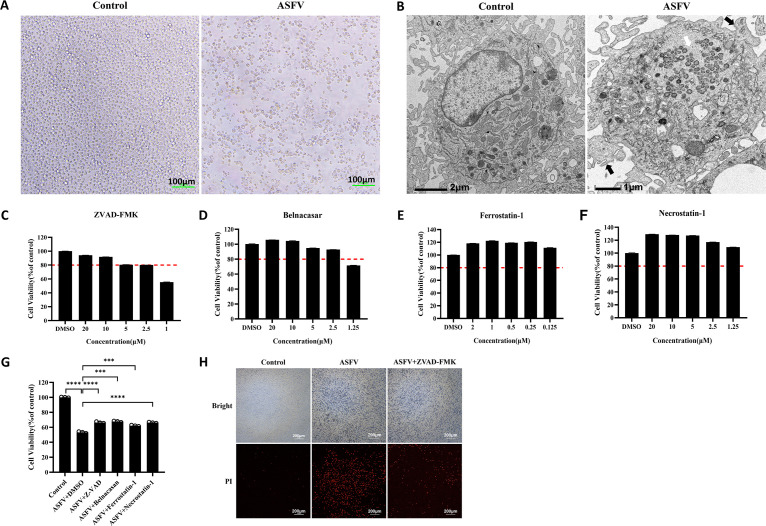
ASF is a severe hemorrhagic disease in pigs characterized by massive depletion and death of infected cells (24). However, the mechanism of cell death induced by ASFV has been poorly studied. PAMs infected with the ASFV GZ201801 strain, with a multiplicity of infection (MOI) of 1, were observed under a microscope, which revealed cytoplasmic vacuolation and cell size reduction with severe cytopathic effects (CPEs) (Fig. 1A). Transmission electron microscope (TEM) also revealed that ASFV-infected PAMs had pathological cells with reduced cell size, increased cytoplasmic density and nucleus concentration, fragmented nucleolus and nuclear membrane, vesicular formation from the cell membrane, and intact cell membrane structure (Fig. 1B). To investigate the way in which ASFV induces cell death, the cytotoxicity of z-VAD-FMK (a pan-caspase inhibitor, used as an apoptosis inhibitor), Belnacasan (selective inhibitor of caspase-1, used as a pyroptosis inhibitor), ferrostatin-1 (a ferroptosis inhibitor), and necrostatin-1 (a cell necrosis inhibitor) was examined in PAMs. The optimal concentrations of 2.5 μM z-VAD-FMK, 2.5 μM Belnacasan, 0.125 μM ferrostatin-1, and 1.25 μM necrostatin-1 were selected for subsequent experiments (Fig. 1C to F). PAMs were infected with 1 MOI of ASFV and incubated with the optimal concentrations of z-VAD-FMK, Belnacasan, ferrostatin-1, and necrostatin-1, after which cell activity was examined. Compared with the ASFV group, the viability of all the drug-treated PAMs was significantly recovered (Fig. 1G), suggesting that ASFV infection induces cell death in various ways, including apoptosis, pyroptosis, ferroptosis, and cell necrosis. To further verify the ASFV-induced CPE-associated cell death pathway in PAMs, the ASFV-infected PAMs were treated with z-VAD-FMK and stained with propidium iodide (PI) to determine the number of dead cells. Notably, z-VAD-FMK significantly inhibited ASFV-induced cell death compared with the ASFV-positive group (Fig. 1H). These results further confirmed that ASFV infection induced apoptosis in the infected PAMs.
FIG 1.
Biological phenomena of ASFV-induced apoptosis. (A) Observation of CPE in PAMs infected with ASFV. Scale bar, 100 μm. (B) Projection electron microscopy images of the cell death of PAMs induced by ASFV infection. In the ASFV-infected PAMs, the white arrow represents viral factories, and the black arrows represent apoptotic bodies. (C to F) Cytotoxicity of z-VAD-FMK, Belnacasan, ferrostatin-1, and necrostatin-1 in PAMs. The red dotted line represents the 80% survival rate of cells treated with the drug. (G) The cell viability of ASFV-infected PAMs posttreatment with 2.5 μM z-VAD-FMK, 2.5 μM Belnacasan, 0.125 μM ferrostatin-1, and 1.25 μM necrostatin-1. Data are presented as mean ± SD. Asterisks indicate statistical difference (one-way ANOVA; n = 3; ***, P < 0.001; ****, P < 0.0001). (H) PAMs were stained with propidium iodide to observe the inhibitory effect of z-VAD-FMK on ASFV-induced cell death. ASFV, African swine fever virus (GZ201801 strain); CPEs, cytopathic effects; PAMs, porcine alveolar macrophages; PI, propidium iodide.
ASFV GZ201801 inhibits apoptosis in the early stage and promotes apoptosis in the late stage.
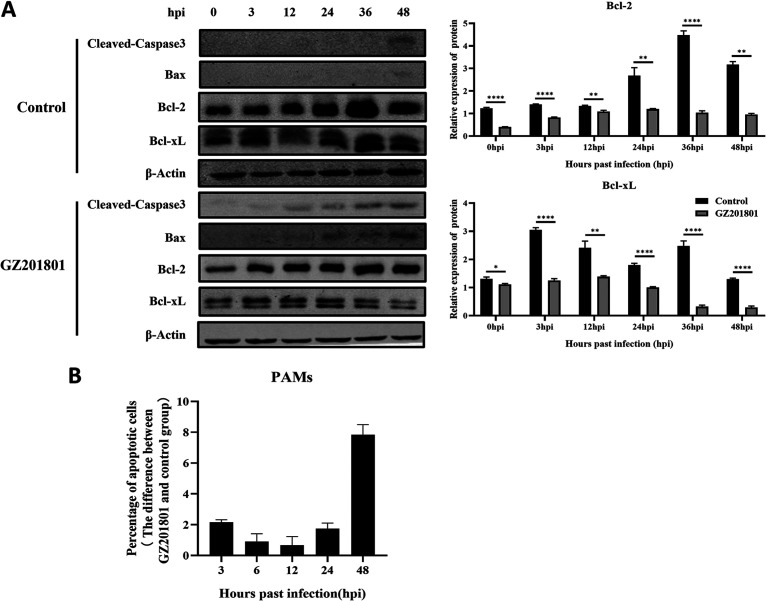
Previous studies have shown that the ASFV GZ201801 strain can induce apoptosis in host cells. Therefore, we examined the expression levels of cleaved-caspase3 protein and proapoptotic protein Bax by Western blotting. ASFV infection increased the expression levels of these two proteins compared with the control group. The expression levels of the antiapoptotic proteins Bcl-2 and Bcl-xL increased initially and then decreased (Fig. 2A). The percentage of apoptosis in the control group and the ASFV-treated group was detected at each time point, and the difference was calculated. Flow cytometry revealed that, compared with that in the control group, the apoptosis of early cells in the ASFV infection group showed a downward trend, while that of late cells showed an upward trend (Fig. 2B). These results indicate that ASFV inhibits apoptosis in the early stages of infection and activates apoptosis in the later stages.
FIG 2.
Characterization of ASFV infection-induced apoptosis of PAMs. (A) Western blotting was used to measure the expression of cleaved-caspase3, Bax, and Bcl-2 proteins in PAMs at 3, 6, 12, 24, 36, and 48 h after infection in the control and ASFV infection groups. β-Actin expression was used as a positive control. (B) Flow cytometry was used to detect apoptosis of PAMs induced by ASFV at 3, 6, 12, 24, and 48 h after infection. ASFV, African swine fever virus (GZ201801 strain); PAMs, porcine alveolar macrophages.
Transcriptomic analysis of PAMs infected with ASFV GZ201801.
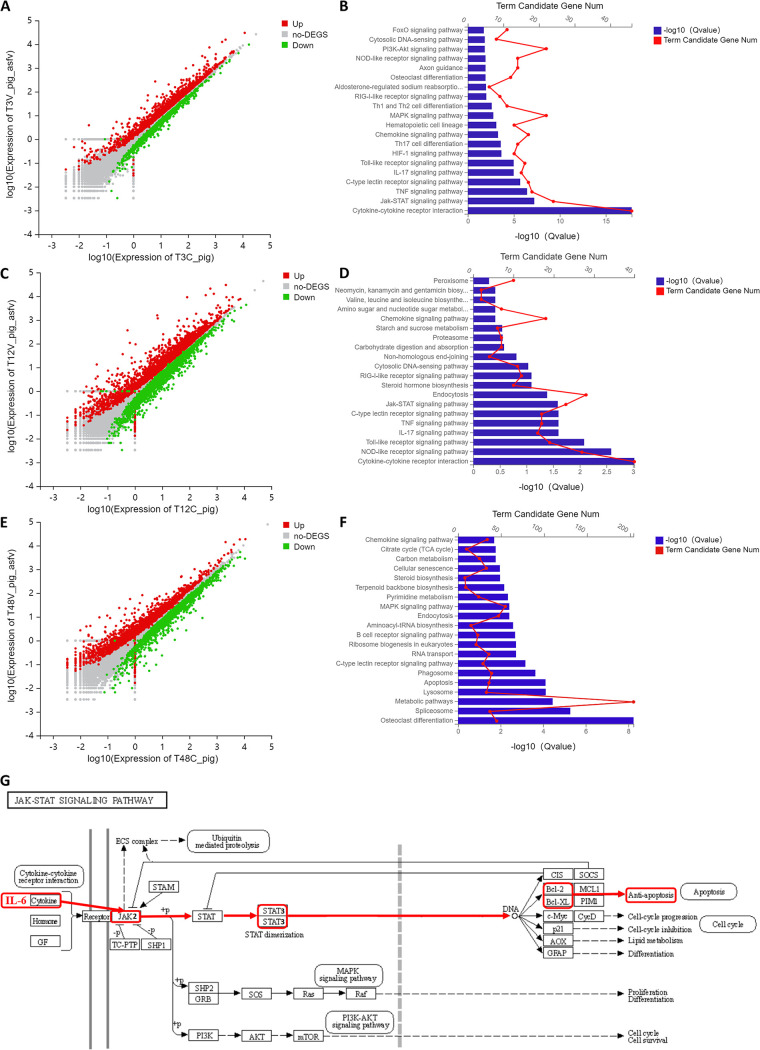
After infection of the PAMs with 1 MOI of ASFV for 3, 12, and 48 h, transcriptomic sequencing was performed. The analysis results showed that, compared with the control group, there were 1,775 upregulated genes and 1,449 downregulated genes after 3 h of infection, 3,023 upregulated genes and 2,657 downregulated genes after 12 h of infection, and 2,087 upregulated genes and 2,056 downregulated genes after 48 h of infection (Fig. 3A, C, and E). KEGG pathway enrichment analysis was performed on the upregulated differential genes at 3, 12, and 48 h after infection. At 3 h after infection, the differentially expressed genes (DEGs) were significantly enriched in the cytokine-cytokine receptor interaction, JAK-STAT signaling pathway, tumor necrosis factor (TNF) signaling pathway, C-type lectin receptor signaling pathway, IL-17 signaling pathway, Toll-like receptor signaling pathway, HIF-1 signaling pathway, Th17 cell differentiation, chemokine signaling pathway, hematopoietic cell lineage, mitogen-activated protein kinase (MAPK) signaling pathway, Th1 and Th2 cell differentiation, and RIG-l-like receptor signaling pathway (Fig. 3B). At 12 h after infection, the DEGs were significantly enriched in the cytokine-cytokine receptor interaction, NOD-like receptor signaling pathway, Toll-like receptor signaling pathway, TNF signaling pathway, IL-17 signaling pathway, C-type lectin receptor signaling pathway, and JAK-STAT signaling pathway (Fig. 3D). At 48 h after infection, the DEGs were significantly enriched in osteoclast differentiation, the spliceosome, metabolic pathways, lysosomes, apoptosis, phagosomes, and C-type lectin receptor signaling pathways (Fig. 3F). These results indicate that ASFV activates the JAK2-STAT3 signaling pathway in the early stage of infection and the apoptosis pathway in the late stage of infection. The KEGG pathway enrichment analysis website (https://www.kegg.jp/dbget-bin/www_bget?map04630) was used to analyze the factors associated with the JAK2-STAT3 pathway. IL-6 cytokines were found to activate the JAK2-STAT3 pathway, promote the phosphorylation of STAT3 in the nucleus, and upregulate the expression of downstream proteins Bcl-2 and Bcl-xL, leading to an antiapoptotic effect (Fig. 3G).
FIG 3.
Transcriptomic analysis of PAMs infected with ASFV. (A, C, and E) Transcriptomic sequencing of PAMs infected with ASFV at MOI to 1. Volcanic map analysis of DEGs compared with that in the control group at 3, 12, and 48 h after infection. (B, D, and F) KEGG pathway enrichment analysis of DEGs compared with the control group at 3, 12, and 48 h after infection. (G) Analysis diagram of the JAK2-STAT3 signaling pathway. ASFV, African swine fever virus (GZ201801 strain); PAMs, porcine alveolar macrophages; MOI, multiplicity of infection; DEGs, differentially expressed genes.
ASFV GZ201801 infection activates the JAK2-STAT3 pathway.
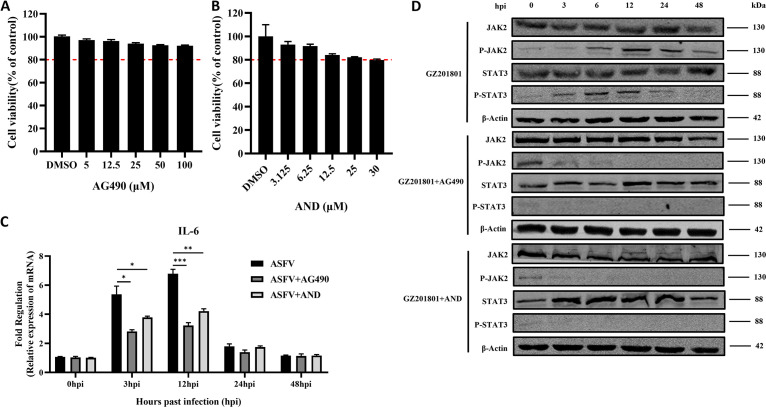
To explore the regulatory effect of ASFV on the JAK2-STAT3 pathway, the cytotoxicity of the JAK2-STAT3 pathway inhibitors AG490 and andrographolide (AND) in PAMs was investigated. The optimal concentrations of AG490 (100 μM) and AND (30 μM) (Fig. 4A and B) were selected for the follow-up experiments. PAMs were infected with ASFV at an MOI of 1 and treated with the optimal concentrations of the pathway inhibitors, after which the expression level of IL-6 was detected by reverse transcriptase quantitative PCR (RT-qPCR). IL-6 was upregulated at the early stage of ASFV infection, and AG490 and AND significantly inhibited the expression of IL-6 (Fig. 4C). Furthermore, Western blotting revealed that the phosphorylation levels of JAK2 and STAT3 proteins were upregulated in the early stages of ASFV infection. In contrast, AG490 and AND downregulated the expression of ASFV infection-induced levels of P-JAK2 and P-STAT3 (Fig. 4D). These results suggest that ASFV infection can promote the expression of IL-6 and activate the JAK2-STAT3 pathway, whereas AG490 and AND can inhibit IL-6 and downregulate the JAK2-STAT3 pathway.
FIG 4.
ASFV infection activates the JAK2-STAT3 pathway. (A and B) Detection of the cytotoxicity of the JAK2-STAT3 pathway inhibitor AG490 (A) and AND (B) in PAMs. The red dotted line represents the 80% survival rate of cells treated with the drug. (C and D) IL-6 expression in PAMs infected with ASFV, with or without treatment with AG490 (C), and Western blotting of the expression of JAK2, P-JAK2, STAT3, and P-STAT3 proteins at 3, 6, 12, 24, 36, and 48 h after infection with ASFV with or without treatment with AG490 and AND (D). β-Actin expression was used as a positive control. ASFV, African swine fever virus (GZ201801 strain); PAMs, porcine alveolar macrophages; AND, andrographolide.
Inhibition of the JAK2-STAT3 pathway promotes ASFV GZ201801-induced apoptosis.
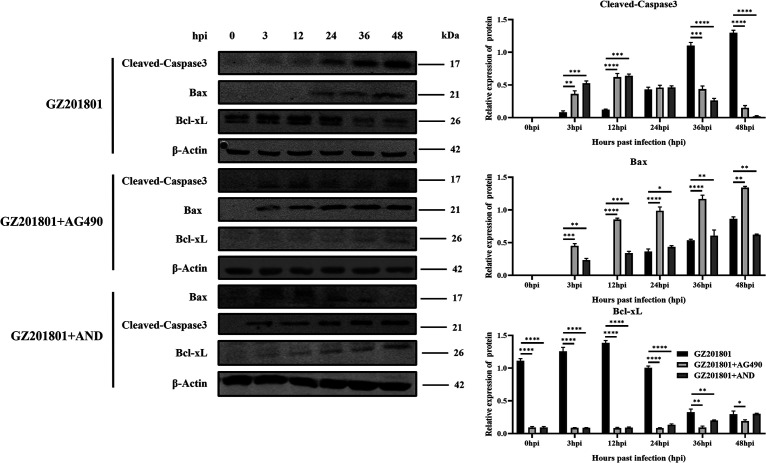
As ASFV infection induces apoptosis, we examined the expression of apoptosis-related proteins at 3, 12, 24, 36, and 48 h in PAMs after infection and with or without treatment with the JAK2-STAT3 pathway inhibitors AG490 and AND by using Western blotting. Upon inhibition of the JAK2-STAT3 pathway, the expression level of cleaved-caspase3 protein and the proapoptotic protein Bax increased in the early stage of ASFV infection. Additionally, the expression level of the antiapoptotic protein Bcl-xL was decreased (Fig. 5). These results suggest that inhibition of the JAK2-STAT3 pathway can significantly promote ASFV-induced apoptosis of PAMs.
FIG 5.
Effect of inhibition of the JAK2-STAT3 pathway on ASFV-induced apoptosis of PAMs. Western blotting of the expression of cleaved-caspase3, Bax, and Bcl-2 proteins at 3, 6, 12, 24, 36, and 48 h after infection with ASFV, with or without treatment with the JAK2-STAT3 pathway inhibitors AG490 and AND. β-Actin expression was used as a positive control. ASFV, African swine fever virus (GZ201801 strain); PAMs, porcine alveolar macrophages; AND, andrographolide; hpi, hours postinfection.
Effects of inhibition of the JAK2-STAT3 pathway on ASFV GZ201801 replication in PAMs.
To explore the effect of the JAK2-STAT3 pathway on the replication ability of ASFV in PAMs, qPCR was used to detect changes in ASFV replication after AG490 and AND treatments. We found that AG490 and AND inhibited the viral load of ASFV (Fig. 6A, D, and G). Furthermore, Western blotting revealed that AG490 and AND could inhibit the expression level of the ASFV p30 protein (Fig. 6B, E, and H). Immunofluorescence assays also indicated that AG490 and AND could significantly inhibit the levels of the ASFV p30 protein (Fig. 6C, F, and I). These results indicate that the replication of ASFV can be inhibited by the suppression of the JAK2-STAT3 pathway by AG490 and AND.
FIG 6.
Antiviral activity of AG490 and AND against ASFV. (A, D, G) PAMs were infected with ASFV (MOI of 1) and then treated with 100 μM AG490 or 30 μM AND. The expression level of the ASFV B646L gene in PAMs was detected by real-time PCR analysis at 48 h. Each data point represents the results of three independent experiments (mean ± SD). (B, E, H) Western blotting of the expression of the ASFV p30 protein in PAMs at 3, 6, 12, 24, 36, and 48 h after infection with ASFV with or without treatment with AG490 and AND. β-Actin expression was used as a positive control. (C, F, I) Antiviral activity of AG490 and AND against ASFV in PAMs determined by IFA. Nuclei were counterstained with DAPI (blue). Each data point represents the results of three independent experiments (mean ± SD). The images represent three independent IFA trials with similar results. Asterisks indicate significant differences compared with the control group (***, P < 0.01). ASFV, African swine fever virus (GZ201801 strain); PAMs, porcine alveolar macrophages; AND, andrographolide; hpi, hours postinfection; IFA, immunofluorescence assay. The same image is used to represent the GZ201801 condition, to better depict and compare the various treatment conditions.
ASFV GZ201801 CD2v induces STAT3 phosphorylation to trigger the JAK2-STAT3 pathway.
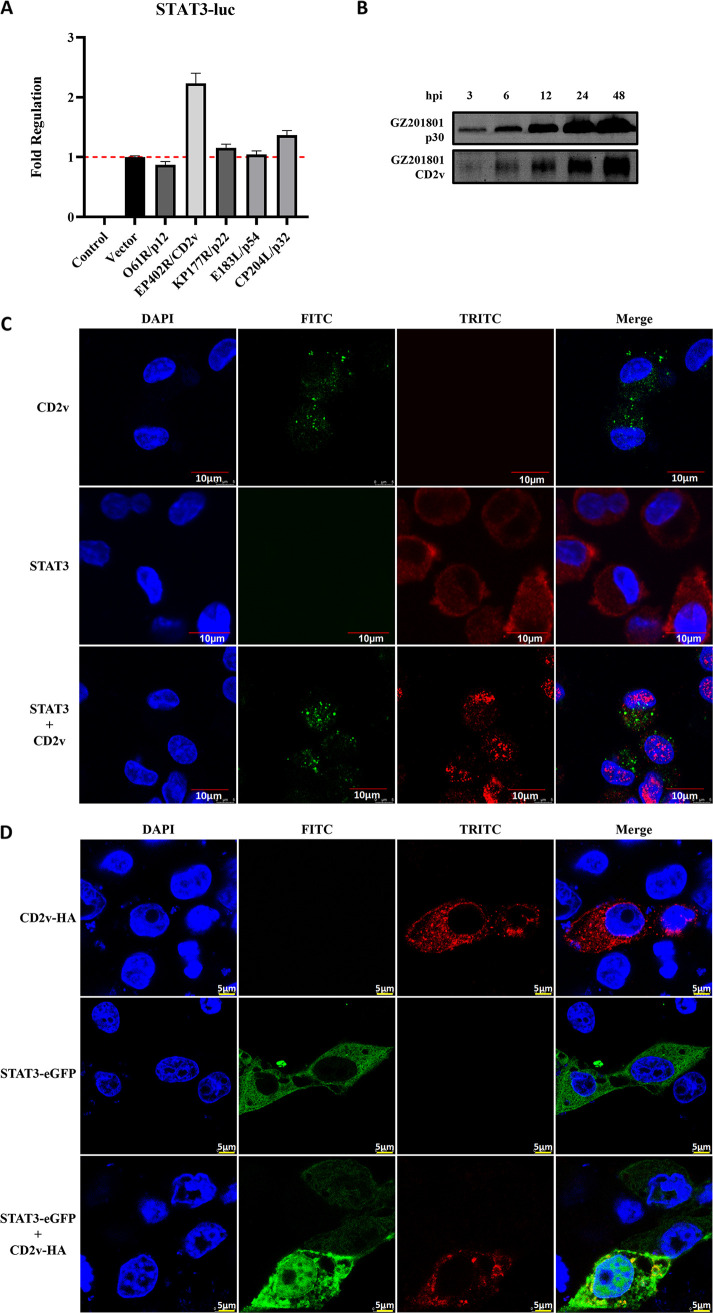
To identify the viral envelope proteins involved in triggering the JAK2-STAT3 pathway, the ASFV envelope proteins p12, CD2v, p22, p54, and p32 were overexpressed in HEK293T cells and screened for their ability to upregulate the STAT3 promoter using the dual-luciferase reporting system. Of these proteins, CD2v showed the most effect (Fig. 7A); hence, it was selected for further examination. The Western blot assay was used to detect the changes in p30 and CD2v protein expression levels during ASFV infection. The results showed that CD2v expression could be detected when PAMs were infected with ASFV for 3 h; however, the expression level was lower than that of p30 (Fig. 7B). Endogenous CD2v and STAT3 proteins were analyzed in PAMs infected with ASFV for 12 h, and nuclear translocation of STAT3 was observed by confocal microscopy after ASFV infection (Fig. 7C). In PK-15 cells, STAT3 was localized in the cytoplasm after 36 h of transfection with the plasmid STAT3 alone. However, upon cotransfection with ASFV CD2v, significant nuclear translocation of the STAT3 protein was observed after 36 h by confocal microscopy (Fig. 7D). These results indicate that ASFV infection can induce the nuclear translocation of STAT3 and that the CD2v protein of ASFV promotes the phosphorylation of STAT3 and its translocation into the nucleus, thus triggering the JAK2-STAT3 pathway.
FIG 7.
ASFV CD2v induces STAT3 phosphorylation and translocation into the nucleus. (A) Screening for capsular proteins that upregulate the STAT3 promoter through the dual luciferase reporting system. The red dotted line is the value of the Vector. (B) Western blot assay was used to detect changes in p30 and CD2v protein expression levels during ASFV infection. (C) Endogenous levels of the CD2v and STAT3 proteins detected in ASFV-infected PAMs, and the nuclear translocation of STAT3 observed through confocal microscopy. (D) The nuclear translocation of STAT3 protein observed by confocal microscopy in PK15 cells after transfection with the plasmid STAT3 alone or STAT3 plasmid + the common ASFV CD2v protein. Cell nuclei were stained with DAPI (blue). ASFV, African swine fever virus (GZ201801 strain); PAMs, porcine alveolar macrophages.
GZ201801-ΔCD2v downregulates the JAK2-STAT3 pathway to promote apoptosis.
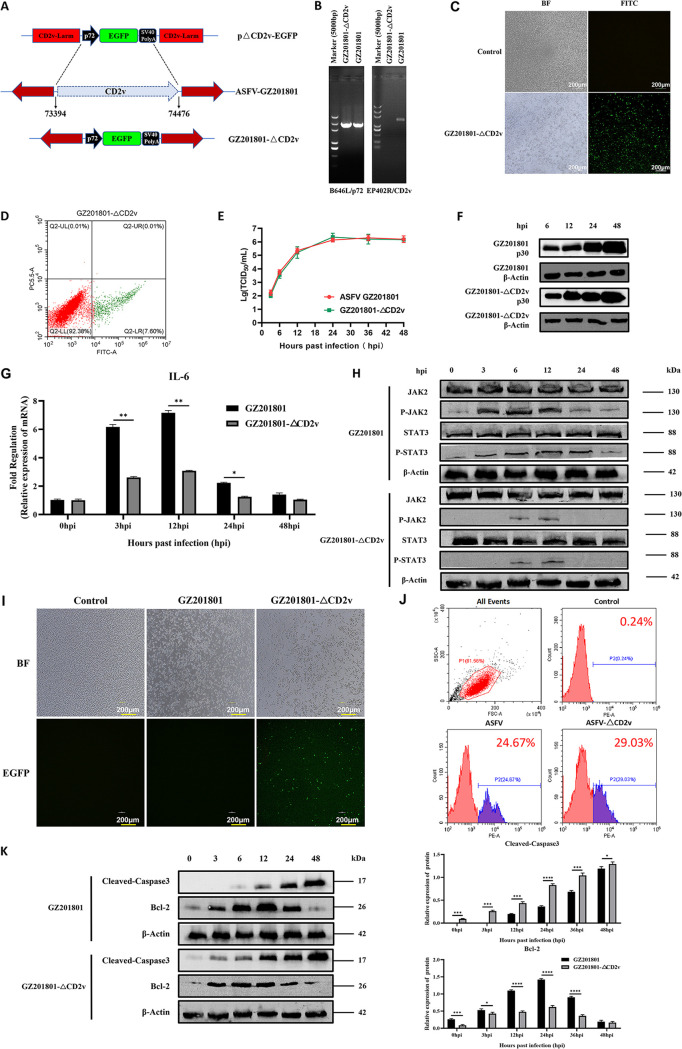
To further verify the regulatory mechanism of ASFV CD2v in the JAK2-STAT3 pathway, the CD2v deletion strain GZ201801-ΔCD2v was first constructed in this study, and its biological characteristics were studied. The EP402R gene expressing CD2v was replaced with the enhanced green fluorescent protein (EGFP) (Fig. 8A) using the homologous recombination method. Meanwhile, the full-length EP402 gene in the GZ201801-ΔCD2v genome was amplified, and the full-length nucleic acid fragment of EP402R could not be amplified (Fig. 8B). The EGFP of the GZ201801-ΔCD2v strain was observed using fluorescence microscopy, and it was found that the deletion virus could express green fluorescence (Fig. 8C). Flow cytometry was used to detect the expression of EGFP of the GZ201801-ΔCD2v strain (Fig. 8D). The above results indicate that the GZ201801-ΔCD2v deletion strain was constructed successfully. Next, the proliferation and growth curves of the GZ201801 and GZ201801-ΔCD2v strains were analyzed, and the proliferation and growth levels of the GZ201801 wild strain and GZ201801-ΔCD2v strain were found to be similar (Fig. 8E). The Western blot assay was used to detect the expression level of the p30 protein in the ASFV-GZ201801 wild strain and GZ201801-ΔCD2v deletion strain during the process of infection in PAMs, and the growth levels of both strains were similar (Fig. 8F). At an MOI of 1, the IL-6 levels of PAMs infected by the GZ201801-ΔCD2v strain were downregulated compared with those in cells infected by the GZ201801 strain (Fig. 8G). Western blotting showed that GZ201801-ΔCD2v inhibited the phosphorylation of JAK2 and STAT3 proteins (Fig. 8H). Microscopy and flow cytometry showed that GZ201801-ΔCD2v infection-induced cell death was higher than that induced by GZ201801 (Fig. 8I and J). Furthermore, Western blotting showed that early GZ201801-ΔCD2v infection could increase the expression level of cleaved-caspase 3 proteins and inhibit the expression of the antiapoptotic Bcl-2 proteins (Fig. 8K).
FIG 8.
GZ201801-ΔCD2v downregulates the JAK2-STAT3 pathway to promote apoptosis. (A) Scheme representing the construction of GZ201801-ΔCD2v. (B) PCR validation of the CD2v deletion. (C and D) Validation of EGFP expression in PAMs infected with the GZ201801-ΔCD2v strain by fluorescence microscopy (C) and flow cytometry (D). The scale bar indicates 200 μm in C. (E) Growth curve replication of the GZ201801 and GZ201801-ΔCD2v strains in PAMs determined with the TCID50 assay (top, primary y axis). (F) Western blot assay used to detect the expression level of the p30 protein in the ASFV-GZ201801 wild strain and GZ201801-ΔCD2v deletion strain during the process of infection in PAMs. (G) Effect of GZ201801 and GZ201801-ΔCD2v infection on IL-6 expression in PAMs. (H) Western blotting of the expression of JAK2, P-JAK2, STAT3, and P-STAT3 proteins at 3, 6, 12, 24, and 48 hpi in GZ201801- and GZ201801-ΔCD2v-infected cells. β-Actin expression was used as a positive control. (I) Cell death induced by GZ201801-ΔCD2v infection observed by microscopy. (J) Number of dead cells post-GZ201801 and GZ201801-ΔCD2v infection detected by flow cytometry (24.67% and 29.03% of PAMs were killed by GZ201801 and GZ201801-ΔCD2v infection, respectively, after 48 h). (K) Western blotting of the expression of cleaved-caspase 3, Bax, and Bcl-2 proteins at 3, 6, 12, 24, 36, and 48 hpi with GZ201801 and GZ201801-ΔCD2v. The data are presented as the mean ± SD (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). GZ201801, wild-type strain of African swine fever virus; GZ201801-ΔCD2v; CD2v deletion strain of GZ201801; PAMs, porcine alveolar macrophages; hpi, hours postinfection. The results shown in H and K were obtained by probing the same membrane with the antibodies mentioned and the membrane was stripped and reprobed with additional antibodies in the figure. As a result, the blots labeled “β-actin” blots in H and K are identical.
ASFV CD2v and CSF2RA interact to regulate the JAK2-STAT3 pathway.
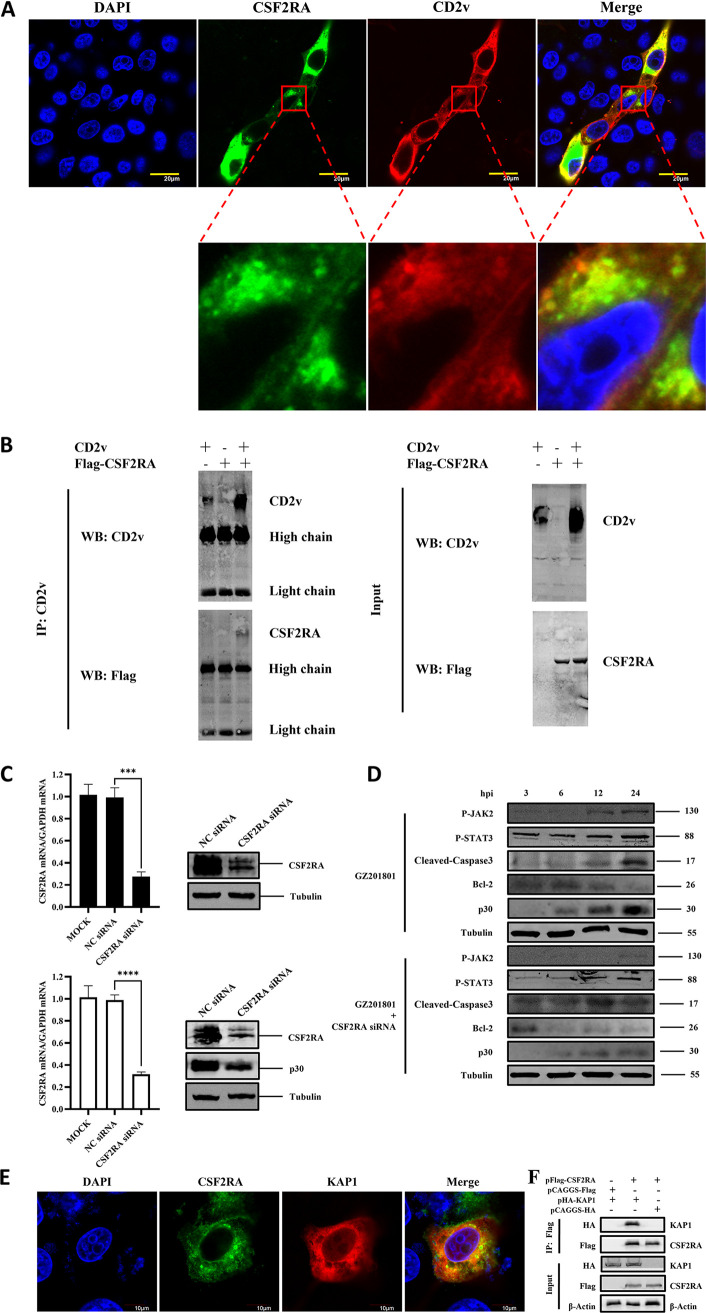
In a previous study, we conducted a yeast two-hybrid assay and revealed that the ASFV CD2v protein potentially interacts with the host protein CSF2RA, which is known to activate the downstream JAK2-STAT3 pathway as a cytokine receptor (26). Therefore, the interaction between ASFV CD2v and CSF2RA was further investigated in this study. The colocalization of CD2v and CSF2RA was observed using confocal microscopy in PK-15 cells (Fig. 9A). Furthermore, an immunoprecipitation assay was performed in HEK-293T cells to evaluate whether CD2v interacts with the CSF2RA complex. As shown in Fig. 9B, CSF2RA coprecipitated with CD2v. To further demonstrate the effect of CSF2RA on CD2v regulation of the JAK2-STAT3 pathway, siRNA was used to interfere with the CSF2RA protein in the host cells (Fig. 9C). Western blot analysis performed on cells infected with ASFV and treated with the CSF2RA siRNA showed that the suppression of CSF2RA expression reduced the ASFV-induced phosphorylation levels of JAK2 and STAT3 proteins, increased the expression of cleaved-caspase3, and inhibited the expression levels of Bcl-2 during early infection. Meanwhile, the expression of the ASFV p30 protein was inhibited (Fig. 9D). These results indicate that CSF2RA interacts with CD2v to activate the JAK2-STAT3 pathway to inhibit apoptosis induced by ASFV in the early stage of infection, thus maintaining viral replication. After siRNA interference of CSF2RA, the JAK2-STAT3 pathway activated early in ASFV infection can be downregulated to promote apoptosis and inhibit viral replication. To further explore how CD2v interacts with CSF2RA to regulate the activation of the JAK2-STAT3 pathway, we studied the interaction between CSF2RA and KAP1. KAP1 is a negative transcriptional regulator of STAT3, and its inhibition can promote the phosphorylation of STAT3 into the nucleus (27). Through confocal microscopy and coimmunoprecipitation experiments, we found that CSF2RA interacts with KAP1 (Fig. 9E and F), suggesting that the activation of the JAK2-STAT3 pathway by ASFV in PAMs is caused by the promotion of the phosphorylation of STAT3 into the nucleus by inhibiting KAP1.
FIG 9.
ASFV CD2v interacts with the host CSF2RA protein to regulate the JAK2-STAT3 pathway. (A) PK-15 cells were cotransfected with HA-CD2v-expressing plasmids and EGFP-CSF2RA-expressing plasmids for 36 h, followed by subcellular localization analysis of ASFV CD2v and CSF2RA by immunofluorescence assay (IFA). Anti-CD2v (red) and anti-CSF2RA (green) antibodies and DAPI (blue) were used to stain the cells. (B) HEK-293T cells were cotransfected with HA-CD2v-expressing plasmids and EGFP-CSF2RA-expressing plasmids for 24 h. The cells were then lysed and subjected to immunoprecipitation (IP) using an anti-CD2v antibody. The IP complexes and WCLs were subjected to Western blotting using anti-CD2v or anti-EGFP antibodies. (C) PAMs were mock transfected or transfected with NC siRNA or CSF2RA siRNA for 24 h, followed by infection with ASFV at MOI of 1 for 24 h. The mRNA expression levels of CSF2RA were then measured by qPCR. The expression levels of CSF2RA and ASFV p30 proteins were detected by Western blotting. (D) Protein expression levels of P-JAK2, P-STAT3, cleaved-caspase3, Bcl-2, and ASFV p30 in PAMs infected with ASFV and treated with CSF2RA siRNA detected by Western blotting. Tubulin expression was used as a positive control. (E) Exploring the interaction between CSF2RA and KAP1 through confocal microscopy. (F) Exploring the interaction between CSF2RA and KAP1 by coimmunoprecipitation. The data are presented as the mean ± SD (n = 3; ***, P < 0.001; ****, P < 0.0001). ASFV, African swine fever virus (GZ201801 strain); PAMs, porcine alveolar macrophages; IP, immunoprecipitation; WCL, whole-cell lysate; NC, Negative Control.
DISCUSSION
In recent years, the rapid spread of ASFV has caused serious economic losses to the global pig industry. An increasing number of studies have shown that ASFV can evade the host immune response through various strategies (28). In this study, we found that early ASFV infection activates the host JAK2-STAT3 signaling pathway to inhibit cell apoptosis and maintain viral replication. The relationship between the virus and host is a dynamic process in which the host immune system tries to resist pathogen infection without damaging itself, and the viruses evolve means of coexisting with the host through immune escape mechanisms. Thus, many viruses establish lifelong infections in their natural hosts, with few clinical manifestations (29). During viral infection, the host cells undergo apoptosis, resulting in the premature death of infected cells and the loss of the replication site of the virus. Therefore, many viruses have developed strategies to prevent apoptosis in infected cells. However, until now, there have been no reports on the inhibition of apoptosis by regulation of the host JAK2-STAT3 pathway in ASFV infection. In this study, based on in vitro screening, ASFV CD2v significantly upregulated the transcription and phosphorylation of STAT3 and its subsequent translocation into the nucleus. Furthermore, CD2v was found to interact with host CSF2RA to activate the JAK2-STAT3 pathway and ultimately inhibit cell apoptosis to maintain viral replication. Thus, our results provide a new direction for further understanding of the immune escape mechanism and pathogenesis of ASFV.
CD2v is an envelope glycoprotein and an important antigenic protein of ASFV. It belongs to the type I transmembrane protein family (30), which is involved in intercellular adhesion, virulence enhancement, and immune response regulation and plays an important role in the pathogenesis of ASFV infection, including tissue adherence, immune escape, and enhanced viral replication (12). CD2v can mediate HAD, induce a protective immune response, participate in immune escape, and enhance viral replication in soft ticks; however, its mechanism is not clear (9, 10, 31, 32). Recently, it has been reported that ASFV K-49 CD2v activates NF-κB, which induces IFN signaling and apoptosis in swine lymphocyte-macrophages (11). However, we found that CD2v inhibited apoptosis in the early phase of infection by regulating the JAK2-STAT3 pathway, which may be due to different results caused by the different strains used. The strain used in the previous report was the virulent ASFV K-49 strain, which is a genotype I and hemagglutination inhibition (HAI) serogroup 2 virus isolated in Congo in 1949 (33). The strain used in our study is the virulent ASFV GZ201801 strain, which is a genotype II and HAI serogroup 8 virus isolated in Guangzhou in 2018. Compared with genotype II ASFV, genotype I ASFV had four large fragment deletions and several base differences. Therefore, our current study is of great significance as it complements the research on apoptosis induced by ASFV strains of different genotypes and serogroups.
In this study, we demonstrated that ASFV infection activated the JAK2-STAT3 pathway and inhibited apoptosis to maintain viral replication. We further demonstrated that the JAK2-STAT3 pathway positively regulates ASFV replication. The JAK2-STAT3 pathway-specific inhibitor AG490 and AND-treated cells significantly blocked JAK2-STAT3 signaling and promoted apoptosis, thus playing an anti-ASFV role. These results confirm that the JAK2-STAT3 pathway is manipulated by ASFV and may be a potential target for inhibiting ASFV infection. Additionally, using a luciferase reporting system, we found that the CD2v protein significantly upregulated the STAT3 transcription level and promoted its phosphorylation and translocation into the nucleus, which prompted us to construct a GZ201801-ΔCD2v deletion strain. Compared with the GZ201801 wild strain, the GZ201801-ΔCD2v deletion strain significantly inhibited the JAK2-STAT3 pathway and promoted apoptosis in host cells at the early stage of infection. These results indicate that CD2v can activate the JAK2-STAT3 pathway to inhibit apoptosis and maintain ASFV replication. To better understand the potential pathogenesis of the CD2v interaction with host proteins, a yeast two-hybrid system was used to screen host proteins for the CD2v interaction, and the interaction between CD2v and CSF2RA was verified using confocal microscopy and coimmunoprecipitation assays. It has been reported that Stat3 activation requires G-CSFR (34), which improves myocardial cell survival after myocardial infarction through JAK2-STAT3 signaling mechanisms (35). In addition, G-CSFR alleviated cardiomyocyte apoptosis after coronary microembolization (CME) in rats by activating the JAK2-STAT3 pathway (29). In this study, we found that interference with siRNA CSF2RA significantly downregulated JAK2-STAT3 signaling and promoted apoptosis, thereby inhibiting ASFV replication.
ASFV is a highly transmissible pathogen that evades host immune responses. Several ASFV proteins, including CD2v, have been shown to inhibit the host antiviral response after interacting with host proteins (36). Here, we found that CD2v interacted with CSF2RA to trigger the JAK2-STAT3 pathway, inhibit apoptosis, and promote ASFV replication. ASFV infection can cause severe inflammatory damage in multiple tissues and organs (37). The imbalance of JAK2-STAT3 signaling induces the body to produce large amounts of interleukin, colony-stimulating factor, and interferon, which may be related to excessive inflammation caused by ASFV (38). Therefore, the CD2v protein may be one of the factors involved in virus-induced diseases.
In this study, the positive regulatory role of the JAK2-STAT3 pathway in ASFV replication was identified. The JAK2-STAT3 signaling triggered during ASFV infection was inhibited by the JAK2-STAT3 pathway inhibitors AG490 and AND to promote cell apoptosis and inhibit viral replication. Thus, the antiviral mechanism underlying the effect of AND was elucidated. In conclusion, this study provides a theoretical basis for the host-immune escape mechanism and pathogenesis of ASFV.
MATERIALS AND METHODS
Cells and viruses.
PAMs are susceptible to ASFV infection and were used for the viral infection experiments in the present study. Primary PAMs were collected from 20- to 30-day-old specific-pathogen-free pigs. The PAMs were maintained in 10% fetal bovine serum + RPMI 1640 medium (Gibco, Waltham, MA) supplemented with 2 mM l-glutamine, 100 U-mL gentamicin, and 0.4 mM nonessential amino acids at 37°C in a 5% CO2 atmosphere saturated with water vapor. PK-15 and HEK-293T cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin and then cultured at 37°C under 5% CO2 conditions. PK-15 and HEK-293T cells are transfected easily and have been used extensively as expression tools for host or viral proteins and to investigate protein-protein interactions. We used HEK-293T cells to perform coimmunoprecipitation assays and identify protein-protein interactions. The highly virulent, hemadsorbing ASFV strain GZ201801 (GenBank MT496893.1), which was isolated in Guangzhou, China, and genotyped as p72 genotype II, and the constructed GZ201801-ΔCD2v strains were preserved at the Infectious Diseases Laboratory of South China Agricultural University.
Reagents and antibodies.
Primers and probes for amplifying genes were synthesized by Invitrogen (Waltham, MA). The following reagents were purchased from the indicated manufacturers: Lipofectamine 2000 (Invitrogen), AceQ universal U+ probe master mix V2 (Vazyme, Nanjing, China), ChamQ universal SYBR qPCR master mix (Vazyme), FastPure viral DNA-RNA minikit (Vazyme), JAK2-STAT3 signal pathway inhibitor AG490 (MedChemExpress, Shanghai, China), Andrographolide (AND; Push Bio-Technology, Chengdu, China), cell counting kit-8 (Beyotime, Shanghai, China), propidium iodide (Beyotime), annexin V-APC apoptosis detection kit (Keygen Biotech, Nanjing, China), one-step PAGE gel fast preparation kit (12%) (Beyotime), bicinchoninic acid (BCA) protein quantification kit (Beyotime), DH5α competent cells (Vazyme), pEASY-basic seamless cloning and assembly kit (Transgen, Beijing, China), DL5000 DNA marker (Vazyme), ultra GelRed (10,000×) (Vazyme), regular agarose (baygene, Shanghai, China), and dual-glo luciferase assay system (Promega, Beijing, China). The murine monoclonal anti-p30 antibody was prepared in our laboratory and used in both Western blotting and immunofluorescence assay (IFAs). Other antibodies and stains were purchased from the indicated manufacturers: anti-α-tubulin rabbit polyclonal antibody (Beyotime), 4′,6-diamidino-2-phenylindole (DAPI; Beyotime), IRDye 800CW goat anti-rabbit IgG antibody, goat anti-mouse IgG antibody (highly cross-adsorbed) (Li-Cor Biosciences, Lincoln, NE), goat anti-rabbit IgG Alexa Fluor 488 and goat anti-mouse IgG Alexa Fluor 594 (Abcam, Cambridge, UK), anti-FLAG M2 Pierce agarose (Sigma, New York, USA), mouse IgG agarose (Beyotime), anti-caspase-3 (D3R6Y) rabbit monoclonal antibody (MAb) (Cell Signaling Technology, Beverly, MA), anti-cleaved-caspase-3 (Asp175) (PDB 5A1E) rabbit MAb (Cell Signaling Technology), anti-JAK2 rabbit monoclonal [EPR108(2)] antibody (Abcam), anti-pJAK2 rabbit monoclonal (E132) antibody (phospho Y1007 + Y1008) (Abcam), anti-STAT3 rabbit monoclonal (EPR787Y) antibody (Abcam), anti-pSTAT3 rabbit monoclonal (EP2147Y) antibody (phospho Y705) (Abcam), anti-Bcl-2 rabbit monoclonal (EPR17509) antibody (Abcam), anti-Bcl-XL rabbit monoclonal (E18) antibody (Abcam), and anti-Bax rabbit monoclonal (E63) antibody (Abcam). siRNA was synthesized by Guangzhou Ribo (CSF2RA siRNA, CGGATCAAAGACAAACTGA).
Viral infection.
PAMs in T25 cell culture flasks were infected with ASFV at an MOI of 1 and incubated for 5 days at 37°C with 5% CO2. Cells and supernatants were harvested and stored at −80°C. For subsequent passaging, 1 mL of the previously passaged virus was used to infect the cells in T25 cell culture flasks. Five successive passages were performed under the same conditions. Following incubation for 2 h at 37°C with 5% CO2, the culture medium was discarded, and the cells were washed twice with phosphate-buffered saline (PBS) and incubated at 37°C with 5% CO2. Cell culture supernatants or cells were collected at 3, 6, 12, 24, 36, and 48 h postinfection (hpi), stored at −80°C, and analyzed to detect extracellular and intracellular virions. Viral genome copy numbers were analyzed using RT-qPCR.
TCID50 assay.
Primary PAMs were cultured in 96-well plates and infected with 10-fold diluted ASFV (GZ201801). IFA experiments were performed on infected PAMs using ASFV p30, and fluorescence conditions were observed and recorded under an inverted fluorescence microscope. The 50% tissue culture infective dose (TCID50) was calculated using the method of Reed and Muench (38, 39). Primary PAMs were infected with ASFV GZ201801 or GZ201801-ΔCD2v strains at an MOI of 1.
RNA isolation, cDNA library preparation, and sequencing.
PAM cells were infected with ASFV (MOI of 1) or blank (control) and harvested at 3, 12, and 48 hpi. Total RNA was extracted from the cells using an RNAiso plus kit (9108; TaKaRa) according to the manufacturer’s instructions. RNA quantity and purity were assessed using a NanoDrop Lite spectrophotometer (ND-NDL-US-CAN; Thermo Fisher Scientific, Waltham, MA). Total RNA was processed using the mRNA enrichment method; mRNA with a polyA tail was enriched with magnetic beads with oligo(dT). Thereafter, database building and sequencing were performed.
Data analysis of transcriptome sequencing (RNA-Seq).
This project used the filtering software SOAPnuke independently developed by BGI for filtering to remove reads containing joints (joint contamination), reads with an unknown base N content greater than 5%, and bases with a mass value of less than 15, which accounts for the total base of the reads for which the proportion of cardinality is greater than 20% of low-quality reads. The raw “reads” were filtered to obtain high-quality de novo transcriptome sequence data.
Plasmid constructs and transfection.
The plasmids pCAGGs-CD2v-HA, pCAGGs-p12-HA, pCAGGs-p22-HA, pCAGGs-p54-HA, pCAGGs-p32-HA, pCAGGs-CSF2RA-FLAG, and pEGFP-C1-STAT3 were constructed using PCR and a pEASY-basic seamless cloning and assembly kit (CU201-02; Transgen, Beijing, China). First, 10 mmol siRNA was added to 50 μL Opti-MEM, and 1.5 μL Lipofectamine 2000 was added to 50 μL Opti-MEM, and placed at 25°C for 5 min. Finally, the two were mixed and placed at 25°C for 20 min. Mixed cultures were added to the complete medium in a 24-well plate. The plates were incubated at 37°C with 5% CO2 for 24 h. Subsequently, viral infection experiments were carried out to test the inhibitory effects.
Cell viability assay.
PAM cells were seeded in a 96-well plate containing maintenance medium and different concentrations of AG490 and AND and incubated at 37°C for 48 h; dimethyl sulfoxide (DMSO) was used as a control (vehicle). Thereafter, 10 μL of the cell counting kit-8 solution was added, and incubation continued for 1 h. After incubation, the absorbance was read at 450 nm using a spectrophotometer, and cell viability was calculated.
Quantitative PCR.
DNA was isolated using the FastPure viral DNA-RNA minikit. A total of 1 μL of DNA was used for the real-time PCR assay using the AceQ universal U+ probe master mix V2. The relative quantity of viral DNA was determined using CADC p72 primers and a probe. The gene-specific primers and probe sequences are listed in Table 1. Total RNA was isolated using the RNAiso plus reagent (9108; TaKaRa) and reverse transcribed into cDNA using the HiScript II 1st strand cDNA synthesis kit (+genomic DNA [gDNA] wiper). A total of 1 μL of cDNA was used for the real-time PCR assay using ChamQ universal SYBR qPCR master mix. The relative quantity of cellular RNA was determined by performing a comparative threshold cycle (ΔΔCT) experiment using GAPDH as an endogenous control. Gene-specific primer sequences were designed using Oligo7 software (Table 1).
TABLE 1.
Primer and probe sequences used in this study for real-time PCR
| Primer | Sequence (5′–3′) |
|---|---|
| B646L | F: ATAGAGATACAGCTCTTCCAG |
| R: GTATGTAAGAGCTGCAGAAC | |
| B646L-probe | FAM-TATCGATAAGATTGAT-MGB |
| IL-6 | F: CCTTCAGTCCAGTCGCCTTCTC |
| R: CATCACCTTTGGCATCTTCTTC | |
| CSF2RA | F: GTCACAAGCCTTGTACTG |
| R: GTCCTCACCTCCTTTTCTGCTG | |
| GAPDH | F: CCTTCCGTGTCCCTACTGCCAAC |
| R: GACGCCTGCTTCACCACCTTCT | |
| CD2v | F: ATGATAATACTTATTTTTTTAATATTTTCTAACATAG |
| R: TTAAATAATTCTATCTACGTGAATAAGCGA | |
| KAP1 | F: ATGGCGGCTTCGGCGGCG |
| R: TCAGGGGCCATCACCGGG |
Western blot analysis.
For Western blot analysis, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (Beyotime) and denatured by adding 4× Laemmli SDS-PAGE buffer (containing DL-dithiothreitol), followed by heating for 15 min at 100°C. The proteins were then separated on SDS-PAGE gels and transferred onto nitrocellulose membranes using a Trans-Blot turbo rapid transfer system (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. The membranes were blocked with 5% defatted milk (dissolved in Tris-buffered saline [TBS]) for 1 h at 37°C and then incubated with a primary antibody for 1 h at 25°C or overnight at 4°C. The membranes were then washed three times (5 min per wash) using a wash buffer (TBS containing 0.1% Tween 20) and incubated with an IRDye 800CW secondary antibody for 1 h at 37°C. The membranes were washed three times in wash buffer and imaged using an Odyssey imaging system (Li-Cor, USA) to visualize the protein bands. α-Tubulin was used as a loading control.
Indirect immunofluorescence assay.
The cells were washed three times with PBS (1 mL each time), fixed in 500 μL 3.7% paraformaldehyde for 30 min at room temperature, and permeabilized in 1 mL 0.1% (wt/vol) Triton-100 for 20 min at room temperature. The cells were incubated with a monoclonal antibody, which was diluted previously with 2% bovine serum albumin (BSA), and then incubated in the dark with a secondary antibody diluted with 2% BSA (1:200) for 1 h at 37°C in a humid chamber. Thereafter, the cell nuclei were stained with 4′,6-diamino-2-phenylindole (DAPI) at room temperature for 5 min and then washed three times with PBS. Cell fluorescence was observed using an immunofluorescence microscope.
Flow cytometry.
After the cells were digested into single cells with 0.25% trypsin, the cells were washed thrice with PBS and stained with the annexin V-APC apoptosis detection kit. An excitation wavelength of 633 nm and a maximum emission wavelength of 660 nm were used to measure cell fluorescence using a Beckman flow cytometer. Analysis was carried out using FlowJo software (v10.8.1; Tree Star, Inc., Ashland, OR).
Coimmunoprecipitation assay.
HEK-293T cells were cultured in 10-cm dishes, and the monolayer cells were cotransfected with the indicated plasmids. The transfected cells were washed with PBS and lysed with 500 μL of lysis buffer. The lysates were subjected to immunoprecipitation, as described previously, using the appropriate antibodies (40). For immunoprecipitation of CD2v with CSF2RA after transfection, cell lysates were immunoprecipitated with anti-CD2v antibody and subjected to Western blotting.
Statistical analysis.
Pathway analysis and functional annotation of the DEGs and differentially expressed proteins identified by transcriptomics and proteomics were performed using KEGG. The SPSS software package (SPSS for Windows v13.0; SPSS Inc., Chicago, IL) was used to perform statistical analysis of the data obtained during the experiment. Differences between the experimental and control groups were analyzed by one-way analysis of variance (ANOVA) using Prism 8 (GraphPad Software, San Diego, CA). Values are expressed in graph bars as the mean ± SD of at least three independent experiments. Statistical significance was set at a P value of <0.05.
ACKNOWLEDGMENTS
This research was funded by the National Key Research and Development Program of China (2021YFD1800100), the Start-up Research Project of Maoming Laboratory (2021TDQD002), Guangzhou Basic and Applied Basic Research Foundation (202201010490), and the China Agriculture Research System of MOF and MARA (cars-35).
Contributor Information
Lang Gong, Email: gonglang@scau.edu.cn.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Sánchez-Vizcaíno JM, Mur L, Martínez-López B. 2013. African swine fever (ASF): five years around Europe. Vet Microbiol 165:45–50. doi: 10.1016/j.vetmic.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 2.Petrov A, Schotte U, Pietschmann J, Dräger C, Beer M, Anheyer-Behmenburg H, Goller KV, Blome S. 2014. Alternative sampling strategies for passive classical and African swine fever surveillance in wild boar. Vet Microbiol 173:360–365. doi: 10.1016/j.vetmic.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 3.Gabriel C, Blome S, Malogolovkin A, Parilov S, Kolbasov D, Teifke JP, Beer M. 2011. Characterization of African swine fever virus Caucasus isolate in European wild boars. Emerg Infect Dis 17:2342–2345. doi: 10.3201/eid1712.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou X, Li N, Luo Y, Liu Y, Miao F, Chen T, Zhang S, Cao P, Li X, Tian K, Qiu HJ, Hu R. 2018. Emergence of African swine fever in China, 2018. Transbound Emerg Dis 65:1482–1484. doi: 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]
- 5.Rowlands RJ, Michaud V, Heath L, Hutchings G, Oura C, Vosloo W, Dwarka R, Onashvili T, Albina E, Dixon LK. 2008. African swine fever virus isolate, Georgia, 2007. Emerg Infect Dis 14:1870–1874. doi: 10.3201/eid1412.080591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malogolovkin A, Burmakina G, Titov I, Sereda A, Gogin A, Baryshnikova E, Kolbasov D. 2015. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg Infect Dis 21:312–315. doi: 10.3201/eid2102.140649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrés G, García-Escudero R, Viñuela E, Salas ML, Rodríguez JM. 2001. African swine fever virus structural protein pE120R is essential for virus transport from assembly sites to plasma membrane but not for infectivity. J Virol 75:6758–6768. doi: 10.1128/JVI.75.15.6758-6768.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alejo A, Matamoros T, Guerra M, Andrés G. 2018. A proteomic atlas of the African swine fever virus particle. J Virol 92:e01293-18. doi: 10.1128/JVI.01293-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borca MV, Kutish GF, Afonso CL, Irusta P, Carrillo C, Brun A, Sussman M, Rock DL. 1994. An African swine fever virus gene with similarity to the T-lymphocyte surface antigen CD2 mediates hemadsorption. Virology 199:463–468. doi: 10.1006/viro.1994.1146. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez JM, Yáñez RJ, Almazán F, Viñuela E, Rodriguez JF. 1993. African swine fever virus encodes a CD2 homolog responsible for the adhesion of erythrocytes to infected cells. J Virol 67:5312–5320. doi: 10.1128/jvi.67.9.5312-5320.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borca MV, Carrillo C, Zsak L, Laegreid WW, Kutish GF, Neilan JG, Burrage TG, Rock DL. 1998. Deletion of a CD2-like gene, 8-DR, from African swine fever virus affects viral infection in domestic swine. J Virol 72:2881–2889. doi: 10.1128/JVI.72.4.2881-2889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowlands RJ, Duarte MM, Boinas F, Hutchings G, Dixon LK. 2009. The CD2v protein enhances African swine fever virus replication in the tick vector, Ornithodoros erraticus. Virology 393:319–328. doi: 10.1016/j.virol.2009.07.040. [DOI] [PubMed] [Google Scholar]
- 13.Korshunova AY, Blagonravov ML, Neborak EV, Syatkin SP, Sklifasovskaya AP, Semyatov SM, Agostinelli E. 2020. BCL2-regulated apoptotic process in myocardial ischemia-reperfusion injury (Review). Int J Mol Med 47:23–36. doi: 10.3892/ijmm.2020.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mima Z, Wang K, Liang M, Wang Y, Liu C, Wei X, Luo F, Nie P, Chen X, Xu Y, Ma Q. 2022. Blockade of JAK2 retards cartilage degeneration and IL-6-induced pain amplification in osteoarthritis. Int Immunopharmacol 113:109340. doi: 10.1016/j.intimp.2022.109340. [DOI] [PubMed] [Google Scholar]
- 15.Zaim Ö, Doğanlar O, Banu Doğanlar Z, Özcan H, Zreigh MM, Kurtdere K. 2022. Novel synthesis naringenin-benzyl piperazine derivatives prevent glioblastoma invasion by inhibiting the hypoxia-induced IL6/JAK2/STAT3 axis and activating caspase-dependent apoptosis. Bioorg Chem 129:106209. doi: 10.1016/j.bioorg.2022.106209. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Prinz RA, Liu X, Xu X. 2020. In vitro and in vivo antiviral activity of gingerenone A on influenza A virus is mediated by targeting Janus kinase 2. Viruses 12:1141. doi: 10.3390/v12101141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Sun J, Hu J, Wang C, Prinz RA, Peng D, Liu X, Xu X. 2020. A77 1726, The active metabolite of the anti-rheumatoid arthritis drug leflunomide, inhibits influenza A virus replication in vitro and in vivo by inhibiting the activity of Janus kinases. FASEB J 34:10132–10145. doi: 10.1096/fj.201902793RR. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Sun J, Prinz RA, Liu X, Xu X. 2020. Inhibition of porcine epidemic diarrhea virus (PEDV) replication by A77 1726 through targeting JAK and Src tyrosine kinases. Virology 551:75–83. doi: 10.1016/j.virol.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, Kanda T, Nakamoto S, Saito K, Nakamura M, Wu S, Haga Y, Sasaki R, Sakamoto N, Shirasawa H, Okamoto H, Yokosuka O. 2015. The JAK2 inhibitor AZD1480 inhibits hepatitis A virus replication in Huh7 cells. Biochem Biophys Res Commun 458:908–912. doi: 10.1016/j.bbrc.2015.02.058. [DOI] [PubMed] [Google Scholar]
- 20.Gajjela BK, Zhou MM. 2022. Calming the cytokine storm of COVID-19 through inhibition of JAK2/STAT3 signaling. Drug Discov Today 27:390–400. doi: 10.1016/j.drudis.2021.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JW, Kuang Y, Chen L, Wang JF. 2018. LncRNA ZNF667-AS1 inhibits inflammatory response and promotes recovery of spinal cord injury via suppressing JAK-STAT pathway. Eur Rev Med Pharmacol Sci 22:7614–7620. doi: 10.26355/eurrev_201811_16375. [DOI] [PubMed] [Google Scholar]
- 22.Fu Y, Zhao Y, Liu Y, Zhu Y, Chi J, Hu J, Zhang X, Yin X. 2012. Adenovirus-mediated tissue factor pathway inhibitor gene transfer induces apoptosis by blocking the phosphorylation of JAK-2/STAT-3 pathway in vascular smooth muscle cells. Cell Signal 24:1909–1917. doi: 10.1016/j.cellsig.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Xu X, Feng X, Zhang B, Wang J. 2011. Adenovirus-mediated delivery of bFGF small interfering RNA reduces STAT3 phosphorylation and induces the depolarization of mitochondria and apoptosis in glioma cells U251. J Exp Clin Cancer Res 30:80. doi: 10.1186/1756-9966-30-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P, Harris D, Liu Z, Rozovski U, Ferrajoli A, Wang Y, Bueso-Ramos C, Hazan-Halevy I, Grgurevic S, Wierda W, Burger J, O'Brien S, Faderl S, Keating M, Estrov Z. 2014. STAT3-activated GM-CSFRα translocates to the nucleus and protects CLL cells from apoptosis. Mol Cancer Res 12:1267–1282. doi: 10.1158/1541-7786.MCR-13-0652-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazan-Halevy I, Harris D, Liu Z, Liu J, Li P, Chen X, Shanker S, Ferrajoli A, Keating MJ, Estrov Z. 2010. STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells. Blood 115:2852–2863. doi: 10.1182/blood-2009-10-230060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li T, Zhao G, Zhang T, Zhang Z, Chen X, Song J, Wang X, Li J, Huang L, Wen L, Li C, Zhao D, He X, Bu Z, Zheng J, Weng C. 2021. African swine fever virus pE199L induces mitochondrial-dependent apoptosis. Viruses 13:2240. doi: 10.3390/v13112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsuruma R, Ohbayashi N, Kamitani S, Ikeda O, Sato N, Muromoto R, Sekine Y, Oritani K, Matsuda T. 2008. Physical and functional interactions between STAT3 and KAP1. Oncogene 27:3054–3059. doi: 10.1038/sj.onc.1210952. [DOI] [PubMed] [Google Scholar]
- 28.Wang W, Ye S, Zhang L, Jiang Q, Chen J, Chen X, Zhang F, Wu H. 2020. Granulocyte colony-stimulating factor attenuates myocardial remodeling and ventricular arrhythmia susceptibility via the JAK2-STAT3 pathway in a rabbit model of coronary microembolization. BMC Cardiovasc Disord 20:85. doi: 10.1186/s12872-020-01385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng X, Nie S, Feng WH. 2022. Regulation of antiviral immune response by African swine fever virus (ASFV). Virol Sin 37:157–167. doi: 10.1016/j.virs.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu XN, Screaton GR, McMichael AJ. 2001. Virus infections: escape, resistance, and counterattack. Immunity 15:867–870. doi: 10.1016/S1074-7613(01)00255-2. [DOI] [PubMed] [Google Scholar]
- 31.Dixon LK, Abrams CC, Bowick G, Goatley LC, Kay-Jackson PC, Chapman D, Liverani E, Nix R, Silk R, Zhang F. 2004. African swine fever virus proteins involved in evading host defence systems. Vet Immunol Immunopathol 100:117–134. doi: 10.1016/j.vetimm.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Chaulagain S, Delhon GA, Khatiwada S, Rock DL. 2021. African swine fever virus CD2v protein induces β-interferon expression and apoptosis in swine peripheral blood mononuclear cells. Viruses 13:1480. doi: 10.3390/v13081480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pérez-Núñez D, García-Urdiales E, Martínez-Bonet M, Nogal ML, Barroso S, Revilla Y, Madrid R. 2015. CD2v interacts with adaptor protein AP-1 during African swine fever infection. PLoS One 10:e0123714. doi: 10.1371/journal.pone.0123714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty A, Dyer KF, Cascio M, Mietzner TA, Tweardy DJ. 1999. Identification of a novel Stat3 recruitment and activation motif within the granulocyte colony-stimulating factor receptor. Blood 93:15–24. doi: 10.1182/blood.V93.1.15. [DOI] [PubMed] [Google Scholar]
- 35.Goatley LC, Dixon LK. 2011. Processing and localization of the African swine fever virus CD2v transmembrane protein. J Virol 85:3294–3305. doi: 10.1128/JVI.01994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou P, Dai J, Zhang K, Wang T, Li LF, Luo Y, Sun Y, Qiu HJ, Li S. 2022. The H240R protein of African swine fever virus inhibits interleukin 1β production by inhibiting NEMO expression and NLRP3 oligomerization. J Virol 96:e0095422. doi: 10.1128/jvi.00954-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lv C, Zhang Q, Zhao L, Yang J, Zou Z, Zhao Y, Li C, Sun X, Lin X, Jin M. 2022. African swine fever virus infection activates inflammatory responses through downregulation of the anti-inflammatory molecule C1QTNF3. Front Immunol 13:1002616. doi: 10.3389/fimmu.2022.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanigur SG, Soydas T, Yenmis G. 2019. NF-κB as the mediator of metformin’s effect on ageing and ageing-related diseases. Clin Exp Pharmacol Physiol 46:413–422. doi: 10.1111/1440-1681.13073. [DOI] [PubMed] [Google Scholar]
- 39.Qin JJ, Cheng X, Zhou F, Lei F, Akolkar G, Cai J, Zhang XJ, Blet A, Xie J, Zhang P, Liu YM, Huang Z, Zhao LP, Lin L, Xia M, Chen MM, Song X, Bai L, Chen Z, Zhang X, Xiang D, Chen J, Xu Q, Ma X, Touyz RM, Gao C, Wang H, Liu L, Mao W, Luo P, Yan Y, Ye P, Chen M, Chen G, Zhu L, She ZG, Huang X, Yuan Y, Zhang BH, Wang Y, Liu PP, Li H. 2020. Redefining cardiac biomarkers in predicting mortality of inpatients with COVID-19. Hypertension 76:1104–1112. doi: 10.1161/HYPERTENSIONAHA.120.15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z, Li C, Du X, Wang G, Cao W, Yang F, Feng H, Zhang X, Shi Z, Liu H, Tian H, Li D, Zhang K, Liu X, Zheng H. 2017. Foot-and-mouth disease virus infection inhibits LGP2 protein expression to exaggerate inflammatory response and promote viral replication. Cell Death Dis 8:e2747. doi: 10.1038/cddis.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]