Abstract
Medical diagnosis assisted by intelligent systems is an effective strategy to increase the efficiency of healthcare systems while reducing their costs. This work is focused on detecting pulmonary conditions from X-ray images using the DeepHealth framework. Our results suggest that it is possible to discriminate pulmonary conditions compatible with the COVID-19 disease from other conditions and healthy individuals. Hence, it could be stated that the DeepHealth framework is a suitable deep-learning software with which to perform reliable medical research. However, more medical data and research are still necessary to train deep learning models that could be trusted by medical personnel.
This project has received funding from the European Commission - Horizon 2020 (H2020) under the DeepHealth Project (grant agreement no 825111) and the SELENE project (grant agreement no 871467).
You have full access to this open access chapter, Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Medical diagnosis is a highly complex process involving a large number of factors. Due to this complexity, the scientific community working in the health sector focuses its efforts on developing systems to assist medical personnel in order to increase its efficiency. With this goal in mind, this work was focused on detecting pulmonary conditions compatible with the COVID-19 disease using chest X-ray images. It might seem that with the success of PCR tests specifically designed for its detection, there would be no reason to perform an X-ray scan on a potential COVID-19 patient. However, in medical practice, it is common to perform chest X-ray scans on patients presenting respiratory problems, so having an automated system for COVID-19 detection using X-ray images can save healthcare systems a lot of time and money.
The significant variability between patients makes the detection of pulmonary conditions from X-ray images a non-trivial problem. In addition to this, COVID-19 can produce symptoms compatible with other diseases, making their diagnosis even more complex. Nonetheless, with more data and more advanced deep learning models, we are confident that these pathologies can be discriminated with the required robustness so that the models can be deployed in production environments.
The main contributions of this work are the following:
-
Ready-to-use pre-trained models to detect COVID-19 related pulmonary conditions in chest X-Ray images with a balanced accuracy of \(83.5\%\), a precision of 0.90, and a recall of 0.95 in the dataset used in this work.
-
A deep learning approach to address the problem of multi-labeling X-ray images giving a score for each target pulmonary condition.
-
Lungs alignment from the extraction of the region of interest where lungs appear in the X-ray image, i.e., semantic segmentation to delimit the silhouette of the lungs.
This paper is organized as follows: Sect. 2 analyses state of the art for COVID-19 and other pulmonary conditions detection using X-ray images. Section 3 gives an introduction to the DeepHealth framework and its features. Section 4 describes all the parts of our experimentation setup. Section 5 shows the experiments carried out and their results. Section 6 presents our conclusions. Finally, Sect. 7 concludes the paper by giving our future research lines.
2 Related Work
Due to the immense impact of COVID-19 and the necessity of quick and reliable screening methods, many studies have arisen to use X-ray images for providing a fast way to detect infected patients. A lot of deep learning approaches have been developed thanks to the power of CNNs to recognize patterns from the images and use such patterns to make predictions.
In the literature, many works can be found achieving high accuracy in classifying chest X-ray images, usually in two (healthy vs. COVID-19) or three (healthy vs. pneumonia vs. COVID-19) classes classification tasks. In Khan et al. [6], they used a novel architecture called STM-RENet combined with Channel Boosting, using two pre-trained models, to perform the two-class classification task achieving an accuracy of 96.53%. In de Moura et al. [4], some architectures well-known in Computer Vision and pre-trained with Imagenet (DenseNet-121 and DenseNet161; ResNet-18 and ResNet-34; VGG-16 and VGG-19) were used to build classification models for the three-class classification task achieving an accuracy of 97.44%. In Kumar et al. [7], a CNN with inception blocks was applied to solve the three-class classification task achieving an accuracy of 97.6%. The model predictions were analyzed using GRAD-CAM (Selvaraju et al. [13]) to see heat maps on the input images with the most relevant pixels for the model in order to make the prediction. In Bhattacharyya et al. [2], they used first a Conditional GAN to segment the lungs; with the extracted lungs, they applied different feature extractors, a Pre-trained VGG19, and the CV algorithm BRISK. The extracted features were combined and fed to a final Random Forest classifier that makes the final prediction, achieving an accuracy of 96.6% in the three-class classification task. In Nayak et al. [8], they tested different popular CNN architectures (AlexNet, VGG-16, GoogleNet, MobileNet-V2, SqueezeNet, ResNet-34, ResNet-50, and Inception-V) pre-trained with the ImageNet dataset, and concluded that the ResNet-34 was the one achieving the best results with an accuracy of 98.33% in the two-class classification task. In Qi et al. [10], they followed a semi-supervised approach using a Teacher-Student architecture to perform the three-class classification task achieving an accuracy of 93%. The model used had two inputs, where one of them was the original image, and the other was a three-channel image where each channel was a filtered version of the original image with three different filters.
One of the problems of working with medical data is the lack of images. The available datasets are usually small, and it is challenging to train robust models with a good generalization ability. A crucial technique to deal with the lack of data is applying Data Augmentation during the training phase. In the works analyzed here, the common practice is to apply affine transformations like Rotation, Flip, Scaling, and Shear. Furthermore, in some cases, other transformations are also applied: Gaussian Noise in Nayak et al. [8]; Elastic Transform, CutBlur, MotionBlur, Intensity Shift, and CutNoise in Kumar et al. [7].
From the analyzed works, it can be seen that researchers usually take more than one dataset to create a more extensive dataset for training; for example, in Nayak et al. [8], they picked the COVID-19 images from one dataset and the healthy samples from another one. This kind of combination can introduce a bias that the model can use just for classifying the samples depending on specific characteristics of each dataset and not original medical-related patterns of COVID-19. In Cruz et al. [3], the authors analyze various COVID-19 datasets and discuss the kinds of biases that can be present when using these datasets. We think this practice is hazardous in these kinds of use cases and even more when the samples of each class come from different datasets.
3 The DeepHealth Framework
The DeepHealth Framework is a flexible and scalable framework for running on HPC and Big Data environments based on two core technology libraries developed within the DeepHealth project: the European Distributed Deep Learning Library (EDDL) and the European Computer Vision Library (ECVL). These libraries will take full advantage of the current and coming development of HPC systems. They will provide a transparent use of heterogeneous hardware accelerators to optimize the training of predictive models while considering performance and accuracy trade-offs.
Both libraries are being integrated into seven software platforms with the DeepHealth project. Similarly, other commercial or research software platforms will have the possibility of integrating the two libraries, given that both are delivered as free and open-source software. Additionally, European large industry companies and SMEs will have the chance of using the DeepHealth Framework for training predictive models on Hybrid HPC + Big Data architectures and export the trained networks using the standard ONNX format [1], so, if their software applications already integrated another DL library able to read the ONNX format, all work is done. The integration of ECVL and EDDL makes it a robust framework with the ability to deal with real-world medical problems by building state-of-the-art models. While ECVL provides support for medical image formats such as DICOM or NIfTI, the EDDL gives the possibility to conduct experiments with modern neural networks on different hardware devices and in a single machine or a distributed environment.
The training pipeline developed in this work uses ECVL to load the images, apply some data augmentation techniques and create the training batches. The EDDL is used to build and train the deep learning models, using the cuDNN implementation as the backend. In addition, two external libraries were used: scikit-image [14] was used to apply the image normalization in the preprocessing step, and scikit-learn [9] to compute some evaluation metrics. The code is available on GitHub (https://github.com/deephealthproject/UC15_pipeline).
4 Experimental Setup
4.1 Datasets
For the COVID-19 classification task, we used the BIMCV-COVID19 dataset [5]. The dataset is divided into two splits, the BIMCV-COVID19+, which contains the data of COVID-19 patients, and the BIMCV-COVID19−, which contains the data of non-COVID-19 patients. For each patient in the dataset, we have one or more sessions available, and each can have one or more X-ray images. Each patient has a list of pathologies that can be used as labels. There are 190 different labels, but the distribution is very unbalanced. We considered just four labels because of their popularity and relation with COVID-19: “covid19”, “normal”, “pneumonia” and “infiltrates”. We prepared two classification tasks, one for binary classification between “normal” and “covid19”, and another for multi-label classification with the four labels (each patient has more than one label, e.g., it can have “infiltrates” and “pneumonia”). From all the available images, we selected just the ones with Anterior-Posterior or Posterior-Anterior views, leaving the ones from the side. We collected 2,083 samples and divided them into three partitions: 1,667 for training, 208 for validation, and 208 for testing. The partition was made at a patient level to avoid having images from the same patient in different splits. The distribution of labels inside each split is shown in Table 1.
4.2 Training Details
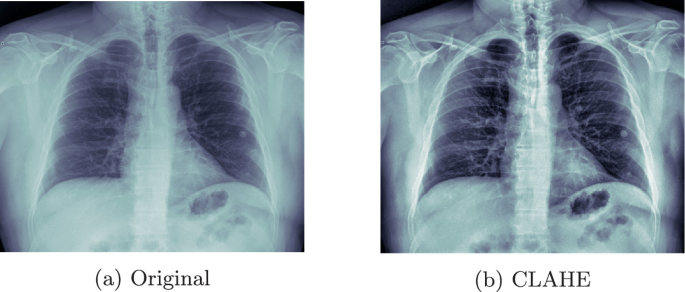
The training images were padded and resized to \(512 \times 512\) and \(256 \times 256\), as the original size was too large to manage (ranging from 2500 to 4000px, height and width). Then their contrast was improved by applying the Contrast Limited Adaptive Histogram Equalization (CLAHE) algorithm (See Fig. 1).
To parallelize the loading of the training images and perform the data augmentation on the fly, the ECVL library was used. The augmentations applied were: Horizontal mirroring, Rotation (from −15 to 15 \(^\circ \)C), Brightness (beta from 0 to 70), and GammaContrast (gamma from 0.6 to 1.4).
Multiple neural network topologies were tested for this task. Specifically, we made use of Imagenet pre-trained models such as VGG-{16, 19}, VGG-{16, 19} with Batch Normalization and ResNet-{18, 50, 101, 152}. These models were downloaded from the ONNX hub and loaded into the EDDL library for training and inference. From each neural model, we added on its top a new classifier with a dense layer with half the number of input features as units, followed by a ReLu activation, a DropOut with a drop rate of 0.4, and finally, a dense layer with Softmax activation to perform the classification. Experiments have shown that pre-trained weights improve training performance and stability.
Next, the weights of the base model (pre-trained feature extractor) were frozen so that the new extension on top of it could be trained for 50 epochs. These were the necessary number of iterations to determine where the loss function became flat. Then, we fine-tuned the entire model with a very small learning rate (1e−5) using SGD for another 50 epochs. To avoid overfitting during training, we applied weight decay (L2) with a penalty of 1e−5, besides the data augmentation and the dropout layers.
All models were trained using a machine with one NVIDIA RTX 3090 GPU (24 GB VRAM), an AMD Ryzen 9 5950X CPU, and 32 GB of RAM. By using this configuration and taking as reference the training time of the experiment with \(256\times 256\) size shown in Table 2. The average training time per batch is 0.055 and 0.016 s for inference. As a result, each training epoch, including training and validation, lasts 15 s on average.
4.3 Evaluation Metrics
To evaluate our models, we used the following metrics:
-
Accuracy: Proportion of correct predictions among the total number of cases examined.
-
Precision (P): Fraction of relevant instances among the retrieved instances.
-
Recall (R): Fraction of relevant instances that were retrieved.
-
F1-score (f1): Harmonic mean of the precision and recall.
-
Intersection Over Union (IoU): Estimates how well a predicted mask matches the ground truth.
5 Experimentation
5.1 Lung Alignment
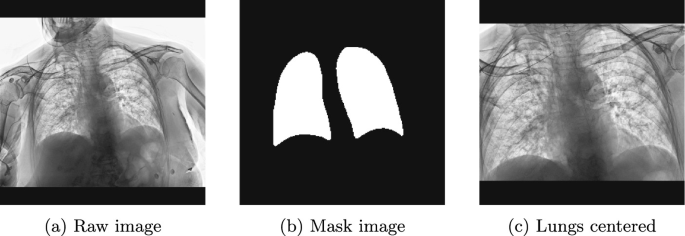
Given the limited memory in our GPUs and the need for using the maximum resolution possible in our images, we decided to crop the lungs regions, excluding all information outside the region of interest (See Fig. 2). To do so, we first segmented the lungs of 250 X-ray images manually, using the tool CVAT [12]. Next, a U-net architecture was used to semi-automatically label the remaining lungs following a human-in-the-loop strategy to reduce human effort, and with which we could efficiently segment the lungs of 2,083 X-ray images, obtaining an IoU of 0.95. That is, we trained a weak segmentation model to segment the non-annotated images (no mask) automatically. By making use of these automatically segmented images, we extended the training set, including the automatic segmentations qualified as correct and discarding those qualified as incorrect. In addition to this, we manually segmented the hard negatives to boost the performance of our model. For the neural architecture, we used the U-Net described in [11] since it is a well-known and widely used neural architecture for image segmentation tasks. As feature extractor we experimented with different models pre-trained on ImageNet such as VGG16, ResNet34, ResNet50, ResNet101, InceptionV3. However, since most of them offered similar results, we finally chose the ResNet34 architecture as it required fewer parameters.
5.2 Detecting Pulmonary Conditions
We experimented with two classification tasks given the selected samples from the BIMCV-COVID19 dataset:
-
Task 1: A binary classification task of “normal” vs “covid19”.
-
Task 2: A multi-label classification task with the four selected labels: “normal”, “covid19”, “pneumonia” and “infiltrates”.
The preprocessing steps, data augmentation, and models used are the same for tasks 1 and 2. The difference appears in the loss functions used and the output layers of the models.
For task 1, the loss functions used are Binary Cross-Entropy (BCE) and regular Cross-Entropy (CE). We used BCE when the label is represented as 0 or 1, and we only used one output neuron in the last layer of the model. The CE was applied when we encoded the labels as one-hot vectors of length two. Regarding task 2, as we are doing a multi-label classification, we used one-hot vectors of length four to encode the labels. In this case, we tried two loss functions: BCE, applying it to each output unit; and Mean Squared Error (MSE).
The best results for the two-class classification task are shown in Table 2. The ResNet101 (pre-trained with Imagenet) was the best model architecture for \(256 \times 256\) and \(512 \times 512\) images. We also observed that increasing the image size does not improve the results; in fact, the best results were obtained with \(256 \times 256\) images.
The best model configuration found in the multi-label task is the \(512 \times 512\) model shown in Table 2. The difference is that the output layer has four neurons and a Sigmoid activation function for the multi-label classification. The loss function used was BCE applied to each output unit independently. The results of this experiment for each class are shown in Table 3. The results obtained indicate that these models are not good enough to be deployed in a production environment. However, the limited amount of training data has to be taken into account since it is to be expected that the accuracy of these models can be significantly improved with more data.
Finally, tabular data (age and sex) was also used to improve the performance of the model. This tabular data was incorporated as a second input connected to a non-linear MLP and concatenated to the final layer of the model. However, no significant results were obtained.
6 Conclusions
This work was focused on detecting pulmonary conditions from X-ray images using the DeepHealth framework.
First, a dataset for lung segmentation was created to extract the regions of interest (lungs) using a U-net model, with the goal of increasing the lungs’ resolution but without increasing their input image size.
Next, we trained several ResNet models (pre-trained with Imagenet) to classify pulmonary conditions, framed as binary or multi-label classification problems, obtaining slightly better results for the binary classification models. Similarly, tabular data (age and sex) was also used to improve the performance of the model, although no significant results were obtained. However, as there are no published results on the BIMCV-COVID19 dataset yet, it is not easy to establish quantitative comparisons with other works. Nonetheless, we would like to highlight the ease of use of the framework for developing advanced deep learning models for medical use cases.
Finally, our work suggests that the DeepHealth Framework is a robust framework to perform reliable medical research. However, given the limited amount of medical data available and the high variability between patients, more research and larger medical datasets are needed before these models can be safely deployed in production environments.
7 Future Work
Many experiments have been left for the future due to time constraints. Nonetheless, we are interested in exploring the following ideas:
-
Can we improve the COVID-19 detection by running a sliding window on a very large image?
-
Can we improve the results by using more X-ray images (from other datasets) to pre-train an autoencoder and then use the encoder part as the backbone for the final classifier model?
References
Bai, J., Lu, F., Zhang, K., et al.: ONNX: Open Neural Network Exchange (2019). https://github.com/onnx/onnx
Bhattacharyya, A., Bhaik, D., Kumar, S., Thakur, P., Sharma, R., Pachori, R.B.: A deep learning based approach for automatic detection of Covid-19 cases using chest X-ray images. Biomed. Signal Process. Control 71, 103182 (2022). https://doi.org/10.1016/j.bspc.2021.103182. https://www.sciencedirect.com/science/article/pii/S1746809421007795
Cruz, B.G.S., Sölter, J., Bossa, M.N., Husch, A.D.: On the composition and limitations of publicly available Covid-19 X-ray imaging datasets. arXiv preprint arXiv:2008.11572 (2020)
de Moura, J., Novo, J., Ortega, M.: Fully automatic deep convolutional approaches for the analysis of Covid-19 using chest X-ray images. Appl. Soft Comput. 115, 108190 (2022). https://doi.org/10.1016/j.asoc.2021.108190. https://www.sciencedirect.com/science/article/pii/S156849462101036X
de la Iglesia Vayá, M., et al.: BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients (2021). https://doi.org/10.21227/w3aw-rv39. https://dx.doi.org/10.21227/w3aw-rv39
Khan, S.H., Sohail, A., Khan, A., Lee, Y.S.: COVID-19 detection in chest X-ray images using a new channel boosted CNN. Diagnostics 12(2) (2022). https://doi.org/10.3390/diagnostics12020267. https://www.mdpi.com/2075-4418/12/2/267
Kumar, A., Tripathi, A.R., Satapathy, S.C., Zhang, Y.D.: SARS-Net: COVID-19 detection from chest X-rays by combining graph convolutional network and convolutional neural network. Pattern Recogn. 122, 108255 (2022). https://doi.org/10.1016/j.patcog.2021.108255. https://www.sciencedirect.com/science/article/pii/S0031320321004350
Nayak, S.R., Nayak, D.R., Sinha, U., Arora, V., Pachori, R.B.: Application of deep learning techniques for detection of COVID-19 cases using chest x-ray images: a comprehensive study. Biomed. Signal Process. Control 64, 102365 (2021). https://doi.org/10.1016/j.bspc.2020.102365. https://www.sciencedirect.com/science/article/pii/S1746809420304717
Pedregosa, F., et al.: Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Qi, X., Foran, D.J., Nosher, J.L., Hacihaliloglu, I.: Multi-feature semi-supervised learning for COVID-19 diagnosis from chest X-ray images. In: Lian, C., Cao, X., Rekik, I., Xu, X., Yan, P. (eds.) MLMI 2021. LNCS, vol. 12966, pp. 151–160. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87589-3_16
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. CoRR abs/1505.04597 (2015). http://arxiv.org/abs/1505.04597
Sekachev, B., et al.: opencv/cvat: v1.1.0, August 2020. https://doi.org/10.5281/zenodo.4009388
Selvaraju, R.R., Das, A., Vedantam, R., Cogswell, M., Parikh, D., Batra, D.: Grad-CAM: why did you say that? Visual explanations from deep networks via gradient-based localization. CoRR abs/1610.02391 (2016). http://arxiv.org/abs/1610.02391
Van der Walt, S., et al.: scikit-image: image processing in Python. PeerJ 2, e453 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Open Access This chapter is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made.
The images or other third party material in this chapter are included in the chapter's Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the chapter's Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
Copyright information
© 2022 The Author(s)
About this paper
Cite this paper
Carrión, S., López-Chilet, Á., Martínez-Bernia, J., Coll-Alonso, J., Chorro-Juan, D., Gómez, J.A. (2022). Detection of Pulmonary Conditions Using the DeepHealth Framework. In: Mazzeo, P.L., Frontoni, E., Sclaroff, S., Distante, C. (eds) Image Analysis and Processing. ICIAP 2022 Workshops. ICIAP 2022. Lecture Notes in Computer Science, vol 13373. Springer, Cham. https://doi.org/10.1007/978-3-031-13321-3_49
Download citation
DOI: https://doi.org/10.1007/978-3-031-13321-3_49
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13320-6
Online ISBN: 978-3-031-13321-3
eBook Packages: Computer ScienceComputer Science (R0)