Abstract
An in-depth insight into the chemistry and nature of the individual chemical bonds is essential for understanding materials. Bonding analysis is thus expected to provide important features for large-scale data analysis and machine learning of material properties. Such chemical bonding information can be computed using the LOBSTER software package, which post-processes modern density functional theory data by projecting the plane wave-based wave functions onto an atomic orbital basis. With the help of a fully automatic workflow, the VASP and LOBSTER software packages are used to generate the data. We then perform bonding analyses on 1520 compounds (insulators and semiconductors) and provide the results as a database. The projected densities of states and bonding indicators are benchmarked on standard density-functional theory computations and available heuristics, respectively. Lastly, we illustrate the predictive power of bonding descriptors by constructing a machine learning model for phononic properties, which shows an increase in prediction accuracies by 27% (mean absolute errors) compared to a benchmark model differing only by not relying on any quantum-chemical bonding features.
Similar content being viewed by others
Background & Summary
Understanding the interactions between constituent atoms in crystalline materials paves the way for developing and tailoring novel solid-state materials with desired application-specific properties1,2,3,4. For instance, the ultra-low lattice thermal conductivity in thermoelectric materials is connected to strong antibonding interactions5,6. Bonding analysis aids in quantifying such interatomic interactions, and several theoretical frameworks exist. Popular and well-known approaches are the Atoms In Molecules (AIM) approach to derive electron density-based Bader charges7, or wave function-based concepts like the Mulliken population analysis8, from which Crystal Orbital Overlap Populations (COOP)9, Crystal Orbital Hamilton Populations (COHP)10, and the Crystal Orbital Bond Index (COBI)11 are derived.
Nowadays, many robust automation frameworks for simulation have become available12,13,14,15,16. These automation tools allow for high-throughput calculations on a scale of thousands of materials17,18,19. Reusing such large amounts of data as inputs for machine learning algorithms has enabled data-driven material science research for accelerated discovery of novel materials and gaining a better understanding between materials structure and properties6,20.
For solid-state materials, plane wave-based basis sets provide easy means to exploit periodicity and gain computational efficiency due to their delocalized nature when performing atomistic simulations via density functional theory (DFT). This computational efficiency comes at the cost of losing crucial atom-specific chemical bonding information. The Local-Orbital Basis Suite Towards Electronic-Structure Reconstruction (LOBSTER)21,22,23,24 software package can recover such bonding information by projecting plane-wave-based wave functions onto atomic orbitals. Since its first release, this program has been used extensively to study different materials classes (e.g, phase-change materials25,26, Li/Na ion battery27,28, low thermal conductivity materials5,29,30,31) and to uncover the diverse underlying atomistic phenomena in the respective bonding mechanisms26,28,31. Although high-throughput materials design and research studies with data have been conducted in a few cases32,33,34,35, no dedicated database exists to retrieve and reuse such data. Previous studies have clearly shown that bonding data computed with LOBSTER is of high value for the materials informatics community, and we provide an open-access database of bonding information here for the first time.
In this work, we perform bonding analysis for 1520 compounds using an automated workflow36 recently developed by some of us that combines Vienna Ab initio Simulation Package (VASP)37,38,39 DFT computations with LOBSTER calculations using Python tools like pymatgen40, atomate14, and FireWorks41. To generate summarized bonding information ready to be used for machine learning studies, we used the LobsterPy36,42 package that automatically analyzes LOBSTER COHP output files. We provide this summarized bonding information data as (lightweight) JSON files. We also distribute all relevant LOBSTER computation data validated and formatted using a pydantic schema, including all the settings and relevant output files.
In the following sections, we begin by briefly summarizing the computational details of the workflow employed to perform the computations. We then describe the method used to generate entries in the database and provide an overview of the structure of the database. Finally, we benchmark the quality of our results by comparing them with projected densities of states from a widely-used density-functional theory code and available heuristics for bond valences and coordination environments. Lastly, we demonstrate the influence of including quantum-chemical bonding data in a machine learning model for predicting phononic properties.
Methods
Structures
We included a total of 1520 crystalline materials in this work. The Materials Project (MP) database43 is used to retrieve all the structures. These materials belong to a previously published dataset of harmonic phonon properties including band structures and densities of states19. We selected this database as it consists only of semiconductors and insulators. For these materials, it is easier to choose a local basis set for the LOBSTER projection as they have clearly distinguished valence and conducting states separated by a band gap. We chose a minimal Slater-type orbital (STO) basis, as provided in LOBSTER, consisting only of occupied valence orbitals in the atomic ground state of each atom (as used in the projector-augmented wave method).
Bonding indicators definitions
LOBSTER first projects the projector-augmented wave (PAW) wavefunctions obtained from DFT computations onto a local STO basis to quantify the interatomic interactions. Combining the coefficients of linear combinations of atomic orbitals (LCAO) generated from this projection with overlap, Hamiltonian, and density matrices, quantum-chemical bonding characteristics in materials are estimated. Here, we summarize the key quantities computed by LOBSTER, and the notations used follow the same convention as in refs. 11,24:
The overlap, Hamiltonian and density matrix between orbitals Φμ and Φv are represented by Sμv, Hμv and Pμv respectively. wk is the k-point weight, and \({c}_{\mu ,jk;\nu ,jk}\) are the coefficients of LCAOs. Re indicates the real part of the complex value. \({\varepsilon }_{j}(k)\) and E represent the energy eigenvalue of band j at k within the Brillouin zone and the general energy, respectively. The energy-integrated values (up to the Fermi level) of these quantities, namely ICOOP, ICOHP, and ICOBI, can be interpreted as the number of electrons in the bond, a measure of bond covalency (corresponding to covalent bond strength), and bond order, respectively.
LOBSTER also provides Mulliken and Löwdin atomic charges from the orbital-derived atomic gross populations (GP)44. The Madelung energy is derived using Mulliken or Löwdin atomic charges as input. Madelung energies represent the electrostatic part of the lattice energy and can be related to the stability of ionic crystal structures. For details about the mathematical formulation related to Madelung energies, Mulliken, and Löwdin atomic charges in LOBSTER, we refer the readers to ref. 11 and the literature referenced therein.
Workflow and computational parameters
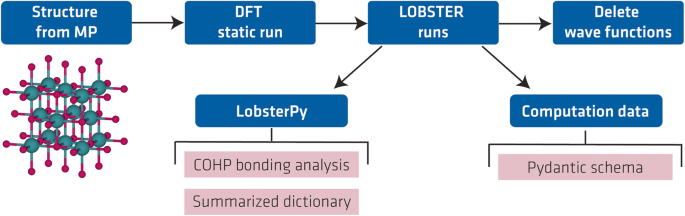
To create the database, we used an automatic bonding analysis workflow36 developed recently by some of us. To start this workflow, one must provide the crystal structure as input. Based on the input structure, it performs the bonding analysis with the LOBSTER24 program by adding all necessary computational steps to the pipeline. To summarize, these steps involve (a) writing VASP input files with an appropriate number of bands (NBANDS) for a static DFT run, (b) a static DFT run, (c) writing input files for LOBSTER runs with all available atomic orbital basis functions for the projection of the wave function, (d) LOBSTER runs, (e) deleting (disk-space consuming) wave function (âWAVECARâ) files. Figure 1 shows the schematic sequence of our workflow.
Within this workflow, the DFT computations were performed using the generalized gradient approximation (GGA) functional as parameterized by Perdew, Burke, and Ernzerhof (PBE)45,46 within the PAW framework47,48. We employ a grid density of 6000 k-points per reciprocal atom and set NEDOS (number of energy points on which the density of states is evaluated) to 10000 points. The electronic structureâs convergence criterion is set to 10â6âeV, and the plane-wave energy cutoff is set to the standard value of 520âeV, as implemented in the original workflow. The Brillouin zone is integrated using the tetrahedron method with Blöchl correction49 (i.e., ISMEARâ=ââ5). All computations were performed including spin polarization. For COHP computations using LOBSTER, we use the entire energy range of VASP static runs, and COHP steps are set equal to the NEDOS (i.e., 10000 steps) set for VASP static run. We stress that the number of COHP steps does not influence the ICOHP values provided in the âICOHPLIST.lobsterâ files; only the energy ranges and increments in the âDOSCAR.lobsterâ and âCO**CAR.lobsterâ are influenced. We increased the number of points for the DOS computation to be able to benchmark the LOBSTER projected DOS with the help of the VASP projected DOS. As both LOBSTER and VASP DOS were computed in the same workflow, the VASP DOS was also computed without symmetry (ISYMâ=â0), which is now also the recommended setting for VASP projected DOS for the VASP version that we used50. With this high number of points in the DOS and COHP computations, the bonding and anti-bonding percentage values from our automatic analysis of output files additionally also pose a very good estimate of bonding and anti-bonding contribution in bonds as we rely on a numerical integration in LobsterPy. All the unprocessed computational data is available (refs. 51,52,53,54,55,56,57,58). The code for starting the workflows is also provided for reproducibility.
Generating data records
We provide data records in two forms. The smaller data record consists of summarized bonding information that is very lightweight and can be quickly assessed in seconds to retrieve and examine relevant bonds. The other, larger data record consists of all the LOBSTER computational data.
To generate the smaller data records including summarized bonding information (LOBSTER lightweight data), we used the âCondensedBondingAnalysisâ schema implemented as part of the atomate259 LOBSTER workflow. This schema automatically analyzes the LOBSTER output files in the âcation-anionâ and âallâ bond modes using the LobsterPy36,42 package. In cases without ions in the structure, only data from the analysis of all bonds are available. When the âcation-anionâ mode is used, the automatic analysis detects cations and anions based on the Mulliken charges, and only âcation-anionâ bonds are included in the analysis. Then, the strongest cation-anion bond is determined based on the Integrated Crystal Orbital Hamilton Populations (ICOHPs). To determine coordination environments and to perform automatic plots, only bonds with a strength of at least 10% of the strongest bond are considered. If the âallâ mode is used, the other bonds are also included in the analysis. The schema also identifies the strongest bonds and corresponding bond lengths based on ICOHP, ICOOP, and ICOBI data for the relevant bond pairs as per LobsterPy bonding analysis. Additionally, we include Madelung energies and atomic charges based on Mulliken and Löwdin population analysis methods. Lastly, a summary of technical validation results, which consists of charge spillings, band overlaps analysis, density of states, and charge comparisons, is included, providing an overview of data quality. A larger data record (Computational data) with all the important LOBSTER computation data is generated using the âLobsterTaskDocumentâ, which is a pydantic schema again implemented as part of the atomate2 LOBSTER workflow. This schema uses LOBSTER parsers implemented in the pymatgen package to read the LOBSTER files and store the information necessary to recreate the Python objects in the form of a Python dictionary. It also includes the LobsterPy data from smaller summarized bonding information data records. A code to generate and read these JSON files is also provided in the code repository for this publication. This allows easy means to reuse or access the data.
Data Records
LOBSTER lightweight data file format
The data is stored in JSON format (ref. 60). The files are named with the the Materials Project ID of the compound. Each JSON file includes summarized bonding information. Table 1 summarizes the root keys to access data from the JSON file. Table 2, explains the data inside the âall_bondsâ and âcation_anion_bondsâ keys. Tables 3, 4 explain the data found in the âlobsterpy_dataâ and âcalc_quality_summaryâ keys of Tables 1, 2, respectively.
Computational data file format
The data is stored in JSON format (refs. 60,61). The files are named as per the Materials Project ID of the compound. Each JSON file includes all the LOBSTER output files parsed and stored in the form of a Python dictionary. It also includes the summarized bonding analysis based on ICOHP values and contains the same information as explained in Table 2. Table 5 summarizes root keys to access data from the JSON file.
Technical Validation
Projection quality
The absolute charge spilling reported at the end of the LOBSTER calculations indicates the quality of the projection corresponding to the loss of charge density that occurs when projecting the original PAW functions onto the local basis. Ideally, when the provided local basis set is complete (i.e., properly reproducing the PAW-based Hilbert space and representing the chemistry of the compound in question), the charge spilling value approaches zero, indicating the reliability of the results. Figure 2 below shows the distribution of the charge spilling for our data set. Approximately 99% of compounds have charge spilling of <5%.
Only a very few compounds show a charge spilling of >5%, possibly due to the limited basis function availability in LOBSTER. The nine compounds showing an absolute charge spilling >5% are BaO2 (mp-1105), SiC (mp-11713), Be2C (mp-1569), Li4NCl (mp-29149), CsBiO2 (mp-29506), Cs2O (mp-7988), KYO2 (mp-8409), Rb2PtSe2 (mp-8622) and SrHfN2 (mp-9383), with spillings ranging between 5.5 and almost 50% (see inset in Fig. 2). The most extreme case is BaO2 with an absolute charge spilling of 46.7%. As the absolute charge spilling determines how well the VASP and LOBSTER wave function match each other for occupied orbitals, two possible reasons for this outlier that are interconnected are coming into consideration: in this particular case, the electronic structure is very sensitive to small changes in the structure and/or the provided basis functions are not sufficient for a proper projection. An additional optimization of the MP structure changes the wave function so that the projection without 5d orbitals arrives at an acceptable absolute charge spilling of 3.91%. Furthermore, with an experimental version of LOBSTER62, that allows to include arbitrary orbitals into the projection, adding the La 5d orbital to Ba, as the VASP POTCAR suggests a 5d occupation of 0.010, the absolute charge spilling drops to 1.40% without further structural optimization. Unfortunately, LOBSTER currently does not provide atomic orbitals for 5d orbitals of Ba.
LOBSTER also generates a âbandOverlaps.lobsterâ file as another measure of projection quality for the cases when the projected wave function is not orthonormalized with an accuracy of 10â5 for every k-point. This file contains the band overlap matrices of the projected bands for each k-point that allows analyzing how well the projected wave function is orthonormalized, and the maximal deviation from the identity matrix is indicated as well. In ideal cases, the deviation should approach zero. Achieving this numerically is almost impossible. Thus, it does not generally indicate a critical error; nevertheless, we analyzed the data from these files for our complete dataset. We set the off-diagonal matrix element deviation threshold for this analysis to 0.1. We then evaluated the percentage of k-points for each compound for which the deviation is larger than the deviation threshold. It is found that approximately 7% of the compounds in the database show 5% or more k-points above this threshold. We have included these compounds in the rest of the analysis and the database, as the other benchmarked results still show sufficient agreement. However, the bonding information from these compounds should be used with caution.
Overall, these results demonstrate that the local basis used for our computations correctly represents the materialâs chemistry for the majority of compounds. The LOBSTER projection mismatch (abs. charge spilling >5%) also helps to figure out problematic basis set functions as discussed in the case of BaO2.
Projected density of states (PDOS) benchmarking
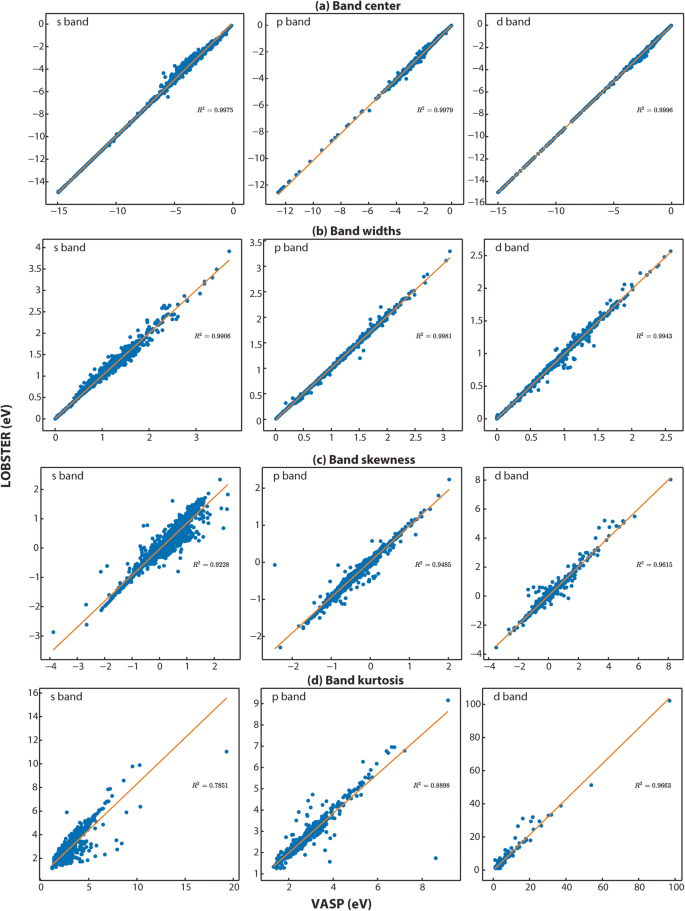
As LOBSTER quantifies the interatomic interactions by projecting the PAW wavefunctions from DFT computations (in our case: VASP) onto a provided local orbital basis, it also generates PDOS that is independent of the PDOS generated by VASP. But unlike the LOBSTER projection, the VASP projection typically loses more electron density when using standard Wigner-Seitz radii. Nevertheless, we will use the VASP projection data for benchmarking as this data is commonly used in the field, and automation are available. We will, however, not compare the absolute projected density of state values for this reason. A common way to compare the density of states relies on visual inspection of relevant features. However, with thousands of PDOS plots, performing a visual inspection is not feasible. To numerically compare the PDOS from VASP and LOBSTER, we have chosen several methods that do not rely on the absolute values but instead on features of the PDOS that are relevant for understanding the electronic structure of a material. First, we compute moments of the PDOS from VASP and LOBSTER. These moments, in principle, provide an estimate of the shape of the PDOS in the selected energy range. Namely, we compare here the band center (1st moment)63, bandwidth (the \(\sqrt{{2}^{nd}}\) moment), band skewness (the 3rd standardized moment), and kurtosis (the 4th standardized moment) of the band directly below the Fermi level (EF). These features provide an overview of the numerical similarity of DOS and are easy to evaluate using existing methods implemented in the âelectronic_structure.dosâ module in pymatgen40,64. It must be noted that we compare the Löwdin symmetric orthonormalized (LSO) DOS obtained from LOBSTER, which recovers the entire Hilbert space and ensures that no electron density is lost, to the VASP projected DOS.
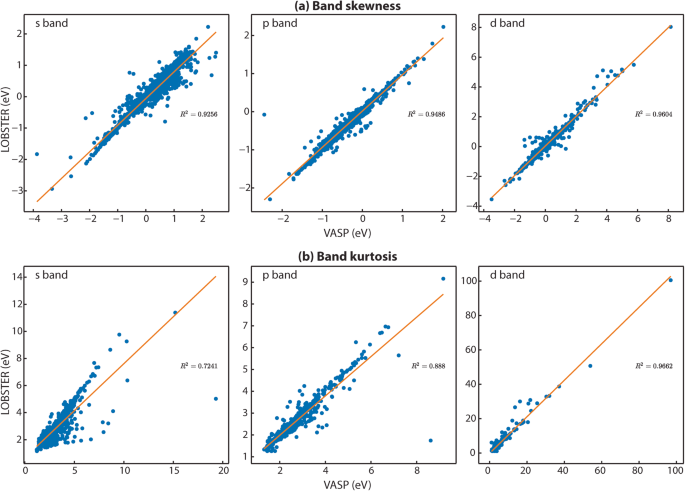
To compute the PDOS features, we first extract all energy ranges below EF in which the PDOS is not equal or close to zero. Next, we use the energy range just below EF, where a non-zero PDOS is detected, to evaluate the PDOS moment features. To ensure that the obtained energy ranges significantly contribute to the overall band, we set a threshold of 0.5 electrons for the band feature comparisons. Figure 3a provides exemplar plots for comparing the PDOS. As evident from the band features, a sufficient agreement exists in this particular case (diamond, mp-66) between VASP and LOBSTER data. In Fig. 4a,b, we compare projected DOS for s, p, and d band centers and band widths obtained from our VASP and LOBSTER runs for the whole data set, respectively. A very good agreement is visible for most compounds. In Fig. 5, we report comparisons of PDOS features, namely band skewness and kurtosis. A comparison of the non-LSO DOS is also shown in the in Fig. 6.
(a) Band features and (b) fingerprint exemplar plots for PDOS from LOBSTER and VASP runs for diamond (mp-66). In subfigure (a), BC, BW, BS, and BK denote band center, width, skewness, and kurtosis, respectively. The percentages of orbital contribution in the chosen energy range are shown in subfigure (b) as % LOBS and %VASP. The Tanimoto index and the normalized vector dot product, respectively, are denoted by the Tanimoto_simi and Norm_simi.
(a) Band centers and (b) band width comparison of projected DOS (s, p and d bands) for the first energy range without PDOS values close or equal to zero below the Fermi level (EF) obtained from LOBSTER and VASP runs. Both figures show that projected DOS from LOBSTER runs agree very well with our reference VASP data. (c) Histogram of Tanimoto index (SA, B) computed between VASP and LOBSTER PDOS (Summed denotes the sum of all individual PDOS).
Another way to assess the similarity between PDOS is to compute Tanimoto coefficients. Earlier studies have demonstrated that such a measure is not only suitable to compute the similarity between molecules65 but is also a reliable way to compare DOS of materials66. The formula to compute the Tanimoto coefficient is as follows:
The Tanimoto coefficient (SA,B) can be interpreted as the ratio of the dot product of the two vectors A and B to the sum of their magnitudes and the dissimilarity between them.
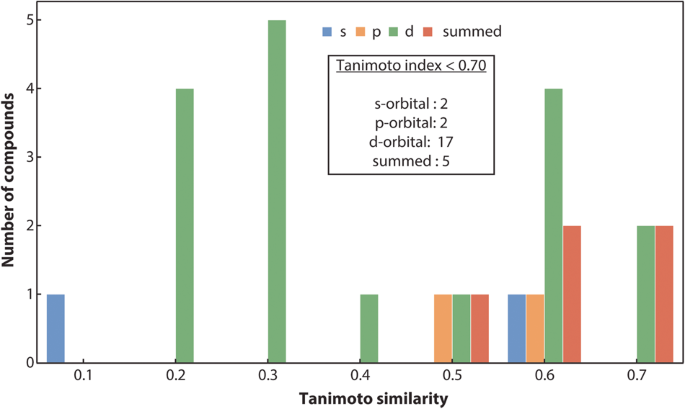
We adapted the âmaterials_fpâ module of the FHI-vibes67,68 Python package to evaluate the similarity between the PDOS of the VASP and the LOBSTER program. The adapted code has been incorporated in the pymatgen package and has been publicly available since v2023.1.9. Here, we first discretize PDOS from VASP and LOBSTER in 256 bins and normalize it before computing the SA, B for the energy range of â15 to 0âeV (energies are shifted relative to the Fermi energy) for all the compounds. Again, for diamond (mp-66) in Fig. 3b,we show the binning of the PDOS and the corresponding Tanimoto similarity, indicating very good agreement between VASP and LOBSTER data. Compounds, where the number of valence electrons obtained by integrating summed PDOS of VASP exceeded the actual valence electrons based on the POTCAR, are excluded from the analysis, as this indicates a poor projection. Again, we only compare PDOS if they significantly contribute to the density of states in the selected energy range. We have set this threshold to 5% of the sum of the projected DOS. Figure 4c shows the distribution of evaluated SA, B for the subset of our dataset. We can see that, for most compounds, SA, B lies in the range of 0.75 to 1. Approximately 99% of compounds have a similarity index of more than 0.70. Only a few cases exist where SA, B is less than 0.70, as shown in Fig. 7. Disagreements are observed in cases where unusual sharp peaks occur in the projection or some low-lying states are missing in the VASP or LOBSTER projections. Overall our results demonstrate that the basic features of the PDOS from VASP and LOBSTER agree very well. Therefore, we can conclude that the LOBSTER projection was performed reliably and that we can compute bonding properties such as COHPs and COBIs of high quality based on this projection. We also provide an interactive dash app to explore these computed PDOS features visually for convenience (https://doi.org/10.5281/zenodo.7795903).
Further quality markers: Atomic charges and coordination environments
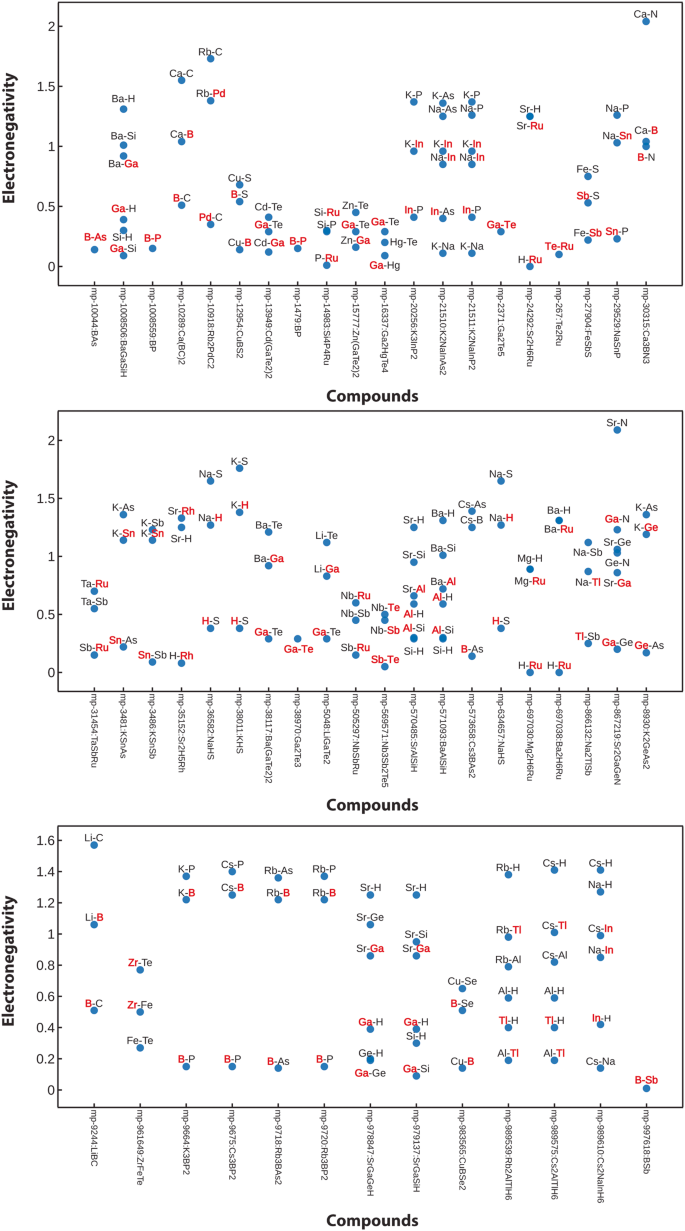
While Mulliken and Löwdin charges from LOBSTER are derived using the LCAO coefficients and arrive at non-integer values44, the bond valence analysis (BVA)69 derives classical integer oxidation states. To make these methods comparable, we chose to sample whether an atomic charge sign from the LOBSTER computations is positive or negative and compare it to the charge signs from the BVA method as implemented in pymatgen. For the two approaches to agree, all constituent atoms in the crystal structure after one-to-one mapping must be classified the same way, i.e., as cations or anions. Here we see 96% agreement between the LOBSTERâs Mulliken charge analysis results and the BVA method. Deviations can be found in compounds having small electronegativity differences between the constituent atom pairs, i.e., for non-ionic compounds. Figure 9 shows the electronegativity difference between atom pairs for compounds where disagreement between BVA and Mulliken atom classification is observed. We highlight the elements where we encounter disagreement in red. A closer look at this figure reveals that a handful of intermetallic, MâH, MâP, and MâB interactions (involving semimetals) are mismatched. An overview of the involved elements is also given as a heatmap in Fig. 8.
Elements for which cations and anions assignment classification differs between LOBSTER and the BVA methods depicted in the form of a heatmap. The heatmap was plotted with pymatviz96.
LobsterPy can evaluate coordination environments directly based on the electronic structure by taking the ICOHP (a covalent bond strength measure) into account36,70,71. The ICOHPs are used to determine the neighboring atoms. In this comparison, we only focus on bonds between cations and anions as determined by the Mulliken charges. Based on the shapes formed by the neighboring atoms, distances to ideal reference polyhedra are then used to determine the closest polyhedra. To validate the coordination environments from LobsterPy, we are benchmarking them with purely geometrically determined ones as determined by ChemEnv70. In ChemEnv, multiple strategies are available to determine coordination environments. Here we use the SimplestChemEnvStrategy to determine the neighbors, which under the hood, uses a Voronoi partitioning scheme. We set the distance and solid-angle cutoffs to recommended values of 1.4 and 0.3, respectively. To only include cation-anion bonds, we again use the BVA method to determine the ideal oxidation states. Comparing the coordination environments detected for each site, we see an agreement for 79% of the sites. Thus, the coordination environments from our database agree very well with those determined by commonly used geometric algorithms.
Data exploration and utility
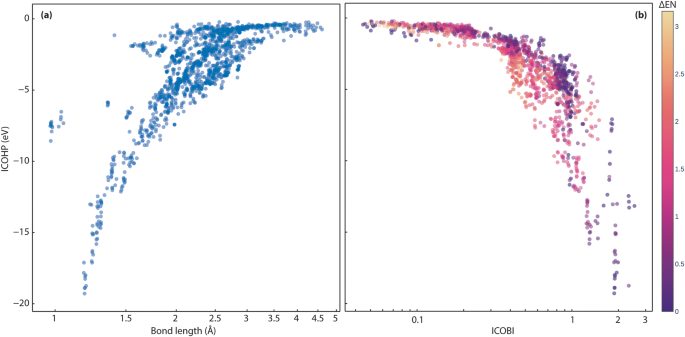
First, we evaluate the bonding indicators in more detail. The most negative ICOHP value indicates the strongest covalent interaction per definition. Plotting the strongest ICOHP values (eV) found per compound and their corresponding bond lengths (à ) as shown in Fig. 10a, we see the expected decrease in covalent bond strengths with increasing bond lengths. In a bond range from about 1âà to 2âà , a steep relation between ICOHP and bond distance can be observed, which eventually flattens for longer bond distances, indicating the short-ranged nature of covalency. The outliers around 1âà within the ICOHP energy range from â5 to â10 eV are OâH and NâH bonds (cf. interactive plots: https://doi.org/10.5281/zenodo.7856484). As covalent bonds between hydrogen and other nonmetal elements are known to be shorter and rather strong in nature72,73,74, this finding is no surprise.
Figure 10b compares the strongest ICOHP and two-center ICOBI interactions for each compound from the LOBSTER computations. Each data point is colored according to the Pauling electronegativity difference (ÎEN) between the interacting atoms. More details can be found in the interactive plot (https://doi.org/10.5281/zenodo.7856484). Up to a bond order (ICOBI) of 0.3 (weak bond range), the change of the ICOHP with growing ICOBI is smaller than after this value. Then, the covalent bond strength increases rapidly with the bond order, demonstrating the different sensitivity of ICOHP and ICOBI with respect to changes in the chemical bonding environment.
Of course, the more ionic interactions (larger ÎEN) can be found within the smaller ICOHP and ICOBI (absolute) values, as both descriptors indicate covalent interactions until eventually only interactions with small ÎEN dominate for the interval ICOHPâ<ââ7 eV and ICOBIâ>â1. The interactions with very small (absolute) ICOHP and ICOBI values labeled as covalent according to ÎEN are metal-metal (weak covalent) interactions like RbâRb or RbâCs contacts. Then there is a range of ICOHP (around â0.7 to â2.0âeV) and ICOBI (around 0.25 to 0.45) values containing Zintl-like intermetallic phases like Na2TlSb (mp-866132), RbAg3Te2 (mp-10481), KZnSb (mp-7438), KCuTe (mp-7436), Na2AgSb (mp-7392), K2AgSb (mp-7643), Na2AgAs (mp-8411), K2CuSb (mp-10381), K5CuSb2 (mp-27999), RbTeAu (mp-9008), K2SbAu (mp-867335), KAuSe2 (mp-29138) or Na2AsAu (mp-7773) and more (ÎEN for the respective bonds ranges between 0.1 and 0.5). This is particularly interesting since Zintl phases and related intermetallic compounds are of great interest for thermoelectric candidates31,75 and, e.g., Na2TlSb76 and KCuTe77 show thermoelectric behavior. Phase-change and thermoelectric materials contain two-center interactions that tend to show smaller ICOHP and ICOBI values than expected from pure electronegativity differences as they are fragments of (hypervalent) multi-center bonds4,11,26,62. In comparison to diamond (ICOHPâ=ââ9.6âeV here and in ref. 4) and silver (ICOHPâ=ââ0.2âeV from ref. 4), the two-center bond characteristic regarding the ICOHP lies between metallic and covalent bonding type (such as GeTe with ICOHPâ=ââ1.8âeV in ref. 4 and ÎENâ=â0.09) and is hence related to the metavalent bonding mechanism26,78,79,80,81. As we have only calculated semiconducting and insulating materials, a purely metallic bonding mechanism can be excluded. Chemically similar compounds in our data set with the classic relation between ICOHP and ÎEN are, e.g., Rb3BaS2 (mp-9718, ICOHP(AsâB)â=ââ7.4âeV, ÎENâ=â0.14), BSb (mp-997618, ICOHPâ=ââ5.0âeV, ÎENâ=â0.01) and Ga2Se3 (mp-1340, ICOHPâ=ââ5.4âeV, ÎENâ=â0.74). It needs to be proven if the relevant compounds from our data set exhibit multi-center ICOBI as well, as it would open up a way to use the ICOHP vs. ICOBI plot as a materials map4,79,80,82,83 for thermoelectric (and phase-change) materials. In summary, we could demonstrate on a larger scale that ICOHP and ICOBI classify bonds according to covalency, and another indicator would be needed to further distinguish the weak covalent interactions as metallic, ionic, or (potential) multi-center interactions.
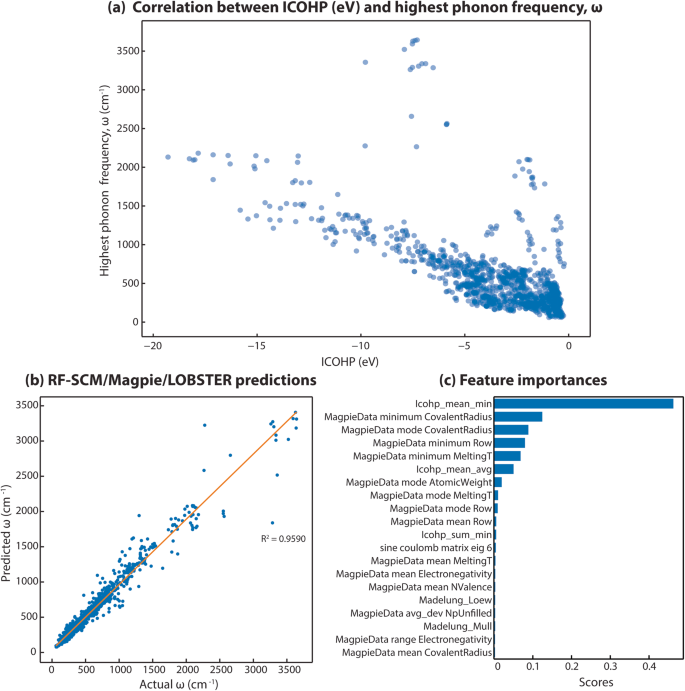
Lastly, we demonstrate the utility of our data by building a machine learning model to predict the highest phonon frequency (Ï) as computed with harmonic phonon computations19. This property is also part of the MatBench benchmark set84. Therefore, a growing number of ML algorithms, such as MegNet85, ALIGNN86, MODNET87,88 have been used to predict the highest phonon frequency. We selected this property as ICOHP values (covalent bond strengths) have previously been correlated to force constants from harmonic phonon runs (e.g., in ref. 89) and should therefore be ideal features for harmonic phonon properties. Also, we have computed LOBSTER data for almost all the compounds included in the benchmark phonon dataset in the MatBench test suit84. We note that bonding analysis only requires a fraction of the computational time of typical phonon runs, as only one static DFT run and post-processing with LOBSTER are required. As a first step before developing the ML model, we checked linear correlations between our quantum-chemical bonding information and our target property. We found a clear correlation between the strongest ICOHP of each compound and the highest phonon frequency (Ï) (Fig. 11a). We can, however, see at least two different trends. We assume this is related to the fact that the highest phonon mode can stem from very different vibrations. Some might be pure stretching vibrations, and others could be collective vibrations involving all atoms. In the first case, mostly one specific bond and one specific ICOHP would have high importance for the phonon mode, whereas in the latter case, all interactions and, therefore, more than one ICOHP within the material would play a role. This observed correlation indicates that using LOBSTER data in ML studies as an additional feature could improve the predictive models.
To test this hypothesis, we first transform the data from summarized bonding information (including all types of bonds and not only cation-anion bonds) of the lightweight JSON files to features for our ML models. For this purpose, we developed a featurizer that accepts these JSON files as input and provides mean, min/max, standard deviation of the ICOHP values, and Madelung energies based on Mulliken and Löwdin as an output in a tabular format for each compound. An explanation of the generated features is provided in Table 6.
Such an approach is commonly used to generate material descriptors for machine learning of material properties87,90. The authors would like to emphasize that the aim of this experiment is not to build the best predictive model but to demonstrate the influence of using LOBSTER data as features in ML studies. We assume that graph-based models which allow adding the bonding descriptors as edge features might be more predictive. That being said, to test the influence on a modelâs predictive performance, we trained and evaluated two Random Forest (RF) regressor91 models. Both models differ only in the input feature sets. RF-SCM/MagPie model consisted of SineCoulombMatrix92 and elemental MagPie90,93 features (mean, average deviation, range, and max/min statistics) as obtained from the âAutoFeaturizerâ module of Automatminer84 with âdebugâ preset (180 features). The input feature set and a fixed set of 500 estimators for RF regressor match the Matbench v0.1 RF-SCM/MagPie model84. The input feature set of the RF-SCM/MagPie/LOBSTER model consisted of the identical feature space as the RF-SCM/MagPie model, and it was augmented by LOBSTER data obtained from our featurizer (199 features). We ensure the train and test sets used for evaluation are identical in both models by setting the same random state seed. The models are evaluated using the nested cross-validation (CV) approach. The inner five-fold CV is used only to optimize the feature selection algorithm (MultiSurfstar94) hyperparameter, i.e., the number of features selected. The hyperparameters of the RF regressor are not tuned. The CV statistics across all five test sets for both models are summarized in Table 7.
Our RF-SCM/Magpie model performs similarly to the one reported on the Matbench test suit84. Including LOBSTER data as features in model input shows an apparent increase in model prediction accuracies. An increase in accuracies by approximately 27% for mean absolute error (MAE), 28% for Max Errors, 32% for root mean squared errors (RMSE), and 5% for R2 is observed.
On further analysis of the best-performing model (RF-SCM/MagPie/LOBSTER), it is found that the algorithm only needs 50 input features after feature selection for predicting the target values more accurately compared to RF-SCM/MagPie, where all 180 were required. This result demonstrates that significantly fewer features are needed when bonding-related features from LOBSTER are included as features. We looked at the feature importance scores readily available for RF models to further analyze the best model. As seen in Fig. 11c, the better performing RF-SCM/MagPie/LOBSTER model shows that the âICOHP_mean_minâ feature, which indicates the ICOHP value for the most covalent bond in a compound, largely contributed to learning the target property of interest. This is the same feature that shows the high correlation in Fig. 11a. Shapley95 values computed for the RF models to assess the impact of input features on model prediction also show a similar trend (plots are provided as part of the repository https://doi.org/10.5281/zenodo.7856481). This result further supports our hypothesis that including bonding-related features as material descriptors in ML studies of materials properties not only improves accuracies of predictions but also helps to understand the relationships between material properties and chemical bonding. Here, we clearly see a suspected relationship between covalent bond strengths and harmonic phonon properties.
Usage Notes
In this work, we provided a Quantum-Chemical Bonding Database to predict and discover new materials. This database consists of summarized COHP-based bonding analysis information ready to be used for ML studies. It also includes (I)COOP, (I)COBI, DOS, atomic charges, and Madelung energies in the computational data JSON files. In addition, we also demonstrated a use-case scenario of how our data could be used for ML studies. This by no means implies that our data should be used in such a manner only. End users are encouraged to explore further.
Code availability
The following program versions have been used in this study: pymatgen 2022.11.7, custodian 2023.3.10, atomate 1.0.3, LOBSTER 4.1.0, and VASP 5.4.4 for VASP and LOBSTER computations using the workflow. For data validation and processing, we have used pymatgen 2023.6.23 and LobsterPy 0.2.9. All the scripts used in this study, from starting the workflow, generating data records, reproducing technical validation plots, and ML model evaluations, can be accessed here: https://github.com/naik-aakash/lobster-database-paper-analysis-scripts (https://doi.org/10.5281/zenodo.8172527).
References
Hoffmann, R. How chemistry and physics meet in the solid state. Angew. Chem. Int. Ed. 26, 846â878, https://doi.org/10.1002/anie.198708461 (1987).
Albright, T. A., Burdett, J. K. & Whangbo, M.-H. Orbital interactions in chemistry, https://doi.org/10.1002/9781118558409 (John Wiley & Sons, 2013).
Burdett, J. K. Chemical bonding in solids (Oxford University Press, 1995).
Chemical bonding with plane waves. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, https://doi.org/10.1016/B978-0-12-823144-9.00120-5 (Elsevier, 2023).
Das, A. et al. Strong antibonding I (p)âCu (d) states lead to intrinsically low thermal conductivity in CuBiI4. J. Am. Chem. Soc. https://doi.org/10.1021/jacs.2c11908 (2023).
He, J. et al. Accelerated discovery and design of ultralow lattice thermal conductivity materials using chemical bonding principles. Adv. Funct. Mater. 32, 2108532, https://doi.org/10.1002/adfm.202108532 (2022).
Bader, R. F. & Nguyen-Dang, T. Quantum theory of atoms in moleculesâdalton revisited. In Advances in Quantum Chemistry, vol. 14, 63â124, https://doi.org/10.1016/S0065-3276(08)60326-3 (Elsevier, 1981).
Mulliken, R. S. Electronic population analysis on LCAOâMO molecular wave functions. I. J. Chem. Phys 23, 1833â1840, https://doi.org/10.1063/1.1740588 (1955).
Hughbanks, T. & Hoffmann, R. Chains of trans-edge-sharing molybdenum octahedra: metal-metal bonding in extended systems. J. Am. Chem. Soc. 105, 3528â3537, https://doi.org/10.1021/ja00349a027 (1983).
Dronskowski, R. & Blöchl, P. E. Crystal orbital hamilton populations (cohp): energy-resolved visualization of chemical bonding in solids based on density-functional calculations. J. Phys. Chem. 97, 8617â8624, https://doi.org/10.1021/j100135a014 (1993).
Müller, P. C., Ertural, C., Hempelmann, J. & Dronskowski, R. Crystal orbital bond index: covalent bond orders in solids. J. Phys. Chem. C 125, 7959â7970, https://doi.org/10.1021/acs.jpcc.1c00718 (2021).
Curtarolo, S. et al. Aflow: An automatic framework for high-throughput materials discovery. Comput. Mater. Sci. 58, 218â226, https://doi.org/10.1016/j.commatsci.2012.02.005 (2012).
Pizzi, G., Cepellotti, A., Sabatini, R., Marzari, N. & Kozinsky, B. Aiida: automated interactive infrastructure and database for computational science. Comput. Mater. Sci. 111, 218â230, https://doi.org/10.1016/j.commatsci.2015.09.013 (2016).
Mathew, K. et al. Atomate: A high-level interface to generate, execute, and analyze computational materials science workflows. Comput. Mater. Sci. 139, 140â152, https://doi.org/10.1016/j.commatsci.2017.07.030 (2017).
Gjerding, M. et al. Atomic simulation recipes: A python framework and library for automated workflows. Comput. Mater. Sci. 199, 110731, https://doi.org/10.1016/j.commatsci.2021.110731 (2021).
George, J. Automation in DFT-based computational materials science. Trends Chem 3, 697â699, https://doi.org/10.1016/j.trechm.2021.07.001 (2021).
Toher, C. et al. High-throughput computational screening of thermal conductivity, Debye temperature, and Grüneisen parameter using a quasiharmonic Debye model. Phys. Rev. B 90, 174107, https://doi.org/10.1103/PhysRevB.90.174107 (2014).
de Jong, M. et al. Charting the complete elastic properties of inorganic crystalline compounds. Sci. Data 2, 150009, https://doi.org/10.1038/sdata.2015.9. Nature Publishing Group (2015).
Petretto, G. et al. High-throughput density-functional perturbation theory phonons for inorganic materials. Sci. Data 5, 1â12, https://doi.org/10.1038/sdata.2018.65 (2018).
Hautier, G. Finding the needle in the haystack: Materials discovery and design through computational ab initio high-throughput screening. Comput. Mater. Sci. 163, 108â116, https://doi.org/10.1016/j.commatsci.2019.02.040 (2019).
Deringer, V. L., Tchougréeff, A. L. & Dronskowski, R. Crystal orbital hamilton population (cohp) analysis as projected from plane-wave basis sets. J. Phys. Chem. A 115, 5461â5466, https://doi.org/10.1021/jp202489s (2011).
Maintz, S., Deringer, V. L., Tchougréeff, A. L. & Dronskowski, R. Analytic projection from plane-wave and paw wavefunctions and application to chemical-bonding analysis in solids. J. Comput. Chem. 34, 2557â2567, https://doi.org/10.1002/jcc.23424 (2013).
Maintz, S., Deringer, V. L., Tchougréeff, A. L. & Dronskowski, R. Lobster: A tool to extract chemical bonding from plane-wave based dft. J. Comput. Chem. 37, 1030â1035, https://doi.org/10.1002/jcc.24300 (2016).
Nelson, R. et al. Lobster: Local orbital projections, atomic charges, and chemical-bonding analysis from projector-augmented-wave-based density-functional theory. J. Comput. Chem. 41, 1931â1940, https://doi.org/10.1002/jcc.26353 (2020).
Konze, P. M., Dronskowski, R. & Deringer, V. L. Exploring chemical bonding in phase-change materials with orbital-based indicators. Phys. Status Solidi - Rapid Res. Lett 13, 1800579, https://doi.org/10.1002/pssr.201800579 (2019).
Hempelmann, J., Müller, P. C., Ertural, C. & Dronskowski, R. The orbital origins of chemical bonding in Ge- Sb- Te phase-change materials. Angew. Chem. Int. Ed. 61, e202115778, https://doi.org/10.1002/anie.202115778 (2022).
Huang, J.-X., Csányi, G., Zhao, J.-B., Cheng, J. & Deringer, V. L. First-principles study of alkali-metal intercalation in disordered carbon anode materials. J. Mater. Chem. A 7, 19070â19080, https://doi.org/10.1039/C9TA05453G (2019).
Ertural, C., Stoffel, R. P., Müller, P. C., Vogt, C. A. & Dronskowski, R. First-principles plane-wave-based exploration of cathode and anode materials for Li-and Na-ion batteries involving complex nitrogen-based anions. chem. Mater. 34, 652â668, https://doi.org/10.1021/acs.chemmater.1c03349 (2022).
Hu, C., Zhou, L., Hu, X., Lv, B. & Gao, Z. Mechanism of the low thermal conductivity in novel two-dimensional NaCuSe. Appl. Surf. Sci. 613, 156064, https://doi.org/10.1016/j.apsusc.2022.156064 (2023).
Dutta, M., Pal, K., Waghmare, U. V. & Biswas, K. Bonding heterogeneity and lone pair induced anharmonicity resulted in ultralow thermal conductivity and promising thermoelectric properties in n-type AgPbBiSe3. Chem. Sci. 10, 4905â4913, https://doi.org/10.1039/C9SC00485H (2019).
Sun, X. et al. Achieving band convergence by tuning the bonding ionicity in n-type Mg3Sb2. J. Comput. Chem. 40, 1693â1700, https://doi.org/10.1002/jcc.25822 (2019).
Xi, L. et al. Discovery of high-performance thermoelectric chalcogenides through reliable high-throughput material screening. J. Am. Chem. Soc. 140, 10785â10793, https://doi.org/10.1021/jacs.8b04704 (2018).
Ohmer, D., Qiang, G., Opahle, I., Singh, H. K. & Zhang, H. High-throughput design of 211- M2AX compounds. Phys. Rev. Mat. 3, 053803, https://doi.org/10.1103/PhysRevMaterials.3.053803 (2019).
Chanussot, L. et al. Open catalyst 2020 (OC20) dataset and community challenges. ACS Catal 11, 6059â6072, https://doi.org/10.1021/acscatal.0c04525 (2021).
Chanussot, L. et al. Correction to âthe open catalyst 2020 (OC20) dataset and community challengesâ. ACS Catal. 11, 13062â13065, https://doi.org/10.1021/acscatal.1c04408 (2021).
George, J. et al. Automated bonding analysis with crystal orbital hamilton populations. ChemPlusChem 87, e202200123, https://doi.org/10.1002/cplu.202200123 (2022).
Kresse, G. & Furthmüller, J. Efficient iterative schemes for ab initio total-energy calculations using a plane-wave basis set. Phys. Rev. B 54, 11169, https://doi.org/10.1103/PhysRevB.54.11169 (1996).
Kresse, G. & Furthmüller, J. Efficiency of ab-initio total energy calculations for metals and semiconductors using a plane-wave basis set. Comput. Mater. Sci. 6, 15â50, https://doi.org/10.1016/0927-0256(96)00008-0 (1996).
Kresse, G. & Hafner, J. Ab initio molecular dynamics for liquid metals. Phys. Rev. B 47, 558, https://doi.org/10.1103/PhysRevB.47.558 (1993).
Ong, S. P. et al. Python materials genomics (pymatgen): A robust, open-source python library for materials analysis. Comput. Mater. Sci. 68, 314â319, https://doi.org/10.1016/j.commatsci.2012.10.028 (2013).
Jain, A. et al. Fireworks: a dynamic workflow system designed for high-throughput applications. Concurr. Comput. Pract.Exper 27, 5037â5059, https://doi.org/10.1002/cpe.3505 (2015).
George, J. et al. Automated Bonding Analysis with Crystal Orbital Hamilton Populations. Zenodo https://doi.org/10.5281/zenodo.7776029 (2023).
Jain, A. et al. Commentary: The materials project: A materials genome approach to accelerating materials innovation. APL Mater 1, 011002, https://doi.org/10.1063/1.4812323 (2013).
Ertural, C., Steinberg, S. & Dronskowski, R. Development of a robust tool to extract mulliken and löwdin charges from plane waves and its application to solid-state materials. RSC Adv 9, 29821â29830, https://doi.org/10.1039/C9RA05190B (2019).
Perdew, J. P. et al. Atoms, molecules, solids, and surfaces: Applications of the generalized gradient approximation for exchange and correlation. Phys. Rev. B 46, 6671, https://doi.org/10.1103/PhysRevB.46.6671 (1992).
Perdew, J. P., Burke, K. & Ernzerhof, M. Generalized gradient approximation made simple. Phys. Rev. Lett. 77, 3865, https://doi.org/10.1103/PhysRevLett.77.3865 (1996).
Kresse, G. & Joubert, D. From ultrasoft pseudopotentials to the projector augmented-wave method. Phys. Rev. B 59, 1758, https://doi.org/10.1103/PhysRevB.59.1758 (1999).
Blöchl, P. E. Projector augmented-wave method. Phys. Rev. B 50, 17953, https://doi.org/10.1103/PhysRevB.50.17953 (1994).
Blöchl, P. E., Jepsen, O. & Andersen, O. K. Improved tetrahedron method for Brillouin-zone integrations. Phys. Rev. B 49, 16223, https://doi.org/10.1103/PhysRevB.49.16223 (1994).
VASP. Lorbit- vaspwiki, https://web.archive.org/web/20230309085254/, https://www.vasp.at/wiki/index.php/LORBIT (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. Quantum-Chemical Bonding Database (Unprocessed data: Part 1). Zenodo https://doi.org/10.5281/zenodo.7852083 (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. Quantum-Chemical Bonding Database (Unprocessed data: Part 2). Zenodo https://doi.org/10.5281/zenodo.7852108 (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. Quantum-Chemical Bonding Database (Unprocessed data: Part 3). Zenodo https://doi.org/10.5281/zenodo.7852792 (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. Quantum-Chemical Bonding Database (Unprocessed data: Part 4). Zenodo https://doi.org/10.5281/zenodo.7852799 (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. Quantum-Chemical Bonding Database (Unprocessed data: Part 5). Zenodo https://doi.org/10.5281/zenodo.7852807 (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. Quantum-Chemical Bonding Database (Unprocessed data: Part 6). Zenodo https://doi.org/10.5281/zenodo.7852809 (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. Quantum-Chemical Bonding Database (Unprocessed data: Part 7). Zenodo https://doi.org/10.5281/zenodo.7852821 (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. Quantum-Chemical Bonding Database (Unprocessed data: Part 8). Zenodo https://doi.org/10.5281/zenodo.7852824 (2023).
Ganose, A. et al. atomate2, https://web.archive.org/web/20230720103837/, https://github.com/materialsproject/atomate2/tree/fa603e3cb4c3024b9b12b0d752793a9191d99f8a (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. A. Quantum-Chemical Bonding Database for Solid- State Materials (JSONS: Part 1). Zenodo https://doi.org/10.5281/zenodo.8091844 (2023).
Naik, A. A., Ertural, C., Dhamrait, N., Benner, P. & George, J. A. Quantum-Chemical Bonding Database for Solid- State Materials (JSONS: Part 2). Zenodo https://doi.org/10.5281/zenodo.8092187 (2023).
Ertural, C. Ãber die elektronische Struktur funktioneller Festkörpermaterialien und ihre Beschreibung mittels lokaler Bindungsindikatoren. Dissertation, RWTH Aachen University https://doi.org/10.18154/RWTH-2022-06735 (2022).
Hammer, B. & Nørskov, J. K. Electronic factors determining the reactivity of metal surfaces. Surf. Sci. 343, 211â220, https://doi.org/10.1016/0039-6028(96)80007-0 (1995).
Rosen, A. S., Vijay, S. & Persson, K. A. Free-atom-like d states beyond the dilute limit of single-atom alloys. Chem. Sci. https://doi.org/10.1039/D2SC05772G (2023).
Bajusz, D., Rácz, A. & Héberger, K. Why is tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminform. 7, 1â13, https://doi.org/10.1186/s13321-015-0069-3 (2015).
Kuban, M., Rigamonti, S., Scheidgen, M. & Draxl, C. Density-of-states similarity descriptor for unsupervised learning from materials data. Sci. Data 9, 646, https://doi.org/10.1038/s41597-022-01754-z (2022).
Knoop, F., Purcell, T., Scheffler, M. & Carbogno, C. Fhi-vibes: Ab initio vibrational simulations. J. Open Source Softw. 5, https://doi.org/10.21105/joss.02671 (2020).
Knoop, F., Purcell, T., Scheffler, M. & Carbogno, C. Fhi-vibes. https://gitlab.com/vibes-developers/vibes/-/tree/master/vibes/materials_fp (2020).
OâKeefe, M. & Brese, N. Atom sizes and bond lengths in molecules and crystals. J. Am. Chem. Soc. 113, 3226â3229, https://doi.org/10.1021/ja00009a002 (1991).
Waroquiers, D. et al. Chemenv: a fast and robust coordination environment identification tool. Acta. Crystallogr. B. 76, 683â695, https://doi.org/10.1107/S2052520620007994 (2020).
Pan, H. et al. Benchmarking coordination number prediction algorithms on inorganic crystal structures. Inorg. Chem. 60, 1590â1603, https://doi.org/10.1021/acs.inorgchem.0c02996 (2021).
Gordy, W. A relation between bond force constants, bond orders, bond lengths, and the electronegativities of the bonded atoms. J. Chem. Phys 14, 305â320, https://doi.org/10.1063/1.1724138 (1946).
Benson, S. W. III-bond energies. J. Chem. Educ. 42, 502â518, https://doi.org/10.1021/ed042p502 (1965).
Missong, R., George, J., Houben, A., Hoelzel, M. & Dronskowski, R. Synthesis, structure, and properties of SrC(NH)3, a nitrogen-based carbonate analogue with the trinacria motif. Angew. Chem. Int. Ed. 54, 12171â12175, https://doi.org/10.1002/anie.201507113 (2015).
Kauzlarich, S. M., Brown, S. R. & Jeffrey Snyder, G. Zintl phases for thermoelectric devices. Dalton Trans. 2099â2107, https://doi.org/10.1039/B702266B (2007).
Yue, T., Zhao, Y., Ni, J., Meng, S. & Dai, Z. Strong quartic anharmonicity, ultralow thermal conductivity, high band degeneracy and good thermoelectric performance in Na2TlSb. Npj Comput. Mater. 9, 17, https://doi.org/10.1038/s41524-023-00970-4 (2023).
Gu, J., Huang, L. & Liu, S. Ultralow lattice thermal conductivity and high thermoelectric performance of monolayer KCuTe: a first principles study. RSC Adv. 9, 36301â36307, https://doi.org/10.1039/C9RA07828B (2019).
Lee, T. H. & Elliott, S. R. Multi-center hyperbonding in phase-change materials. Phys. Status Solidi - Rapid Res. Lett 15, 2000516, https://doi.org/10.1002/pssr.202000516 (2021).
Yu, Y., Cagnoni, M., Cojocaru-Mirédin, O. & Wuttig, M. Chalcogenide thermoelectrics empowered by an unconventional bonding mechanism. Adv. Funct. Mater. 30, 1904862, https://doi.org/10.1002/adfm.201904862 (2020).
Pries, J., Cojocaru-Miredin, O. & Wuttig, M. Phase-change materials: Empowered by an unconventional bonding mechanism. MRS Bulletin 44, 699â704, https://doi.org/10.1557/mrs.2019.204 (2019).
Jones, R. O. The chemical bond in solidsârevisited. J. Condens. Matter Phys 34, 343001, https://doi.org/10.1088/1361-648x/ac7494 (2022).
Esser, M., Maintz, S. & Dronskowski, R. Automated first-principles mapping for phase-change materials. J. Comput.Chem. 38, 620â628, https://doi.org/10.1002/jcc.24724 (2017).
Schön, C.-F. et al. Classification of properties and their relation to chemical bonding: Essential steps toward the inverse design of functional materials. Sci. Adv. 8, eade0828, https://doi.org/10.1126/sciadv.ade0828 (2022).
Dunn, A., Wang, Q., Ganose, A., Dopp, D. & Jain, A. Benchmarking materials property prediction methods: the Matbench test set and Automatminer reference algorithm. Npj Comput. Mater. 6, 138, https://doi.org/10.1038/s41524-020-00406-3 (2020).
Chen, C., Ye, W., Zuo, Y., Zheng, C. & Ong, S. P. Graph networks as a universal machine learning framework for molecules and crystals. Chem. Mater. 31, 3564â3572, https://doi.org/10.1021/acs.chemmater.9b01294 (2019).
Choudhary, K. & DeCost, B. Atomistic line graph neural network for improved materials property predictions. Npj Comput. Mater. 7, 185, https://doi.org/10.1038/s41524-021-00650-1 (2021).
De Breuck, P.-P., Hautier, G. & Rignanese, G.-M. Materials property prediction for limited datasets enabled by feature selection and joint learning with modnet. Npj Comput. Mater. 7, 83, https://doi.org/10.1038/s41524-021-00552-2 (2021).
De Breuck, P.-P., Evans, M. L. & Rignanese, G.-M. Robust model benchmarking and bias-imbalance in data-driven materials science: a case study on modnet. J. Condens. Matter Phys 33, 404002, https://doi.org/10.1088/1361-648X/ac1280 (2021).
Deringer, V. L., Stoffel, R. P., Wuttig, M. & Dronskowski, R. Vibrational properties and bonding nature of Sb2Se3 and their implications for chalcogenide materials. Chem. Sci. 6, 5255â5262, https://doi.org/10.1039/C5SC00825E Royal Society of Chemistry (2015).
Ward, L. et al. Matminer: An open-source toolkit for materials data mining. Comput. Mater. Sci. 152, 60â69, https://doi.org/10.1016/j.commatsci.2018.05.018 (2018).
Breiman, L. Random forests. Machine learning 45, 5â32, https://doi.org/10.1023/A:1010933404324 (2001).
Faber, F., Lindmaa, A., Von Lilienfeld, O. A. & Armiento, R. Crystal structure representations for machine learning models of formation energies. Int. J. Quantum Chem. 115, 1094â1101, https://doi.org/10.1002/qua.24917 (2015).
Ward, L., Agrawal, A., Choudhary, A. & Wolverton, C. A general-purpose machine learning framework for predicting properties of inorganic materials. Npj Comput. Mater. 2, 1â7, https://doi.org/10.1038/npjcompumats.2016.28 (2016).
Urbanowicz, R. J., Olson, R. S., Schmitt, P., Meeker, M. & Moore, J. H. Benchmarking relief-based feature selection methods for bioinformatics data mining. J. Biomed. Inform. 85, 168â188, https://doi.org/10.1016/j.jbi.2018.07.015 (2018).
Lundberg, S. M. et al. From local explanations to global understanding with explainable ai for trees. Nat. Mach. Intell 2, 56â67, https://doi.org/10.1038/s42256-019-0138-9 (2020).
Riebesell, J., Goodall, R. & Baird, S. G. Pymatviz https://doi.org/10.5281/zenodo.7486816 (2022).
Acknowledgements
A.N. and J.G would like to acknowledge the Gauss Centre for Super computing e.V. (www.gauss-centre.eu) for funding this project by providing generous computing time on the GCS Supercomputer SuperMUC-NG at Leibniz Super computing Centre (www.lrz.de) (project pn73da). The authors thank Katharina Ueltzen for bringing to light an issue with supplied magnetic moments from INCAR not being read correctly during VASP static runs and for helping us rectify the same. A.N. thanks Franziska Emmerling and Manuel Kupper for their feedback on the manuscript in BAMâs MatChIngCamp. J.G. thanks Geoffroy Hautier and Matthew Horton for helpful discussions, and A.N. and J.G. thank Alex Ganose for reviewing the pydantic schema used in this study as part of our atomate2 pull request for a new LOBSTER workflow. We also acknowledge the maintainers of pymatgen.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
A.N. performed the high-throughput calculations and data collection with help from J.G. and P.B.C.E. performed additional computations to analyze the BaO2 case. All authors analyzed the data. A.N., C.E. and J.G. wrote the manuscript with inputs from all authors. A.N. and J.G. have planned the study. A.N., N.D., P.B. and J.G. contributed to the ML model.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisherâs note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the articleâs Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the articleâs Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Naik, A.A., Ertural, C., Dhamrait, N. et al. A Quantum-Chemical Bonding Database for Solid-State Materials. Sci Data 10, 610 (2023). https://doi.org/10.1038/s41597-023-02477-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02477-5