Abstract
Pre-trained transformer language models (LMs) on large unlabeled corpus have produced state-of-the-art results in natural language processing, organic molecule design, and protein sequence generation. However, no such models have been applied to learn the composition patterns for the generative design of material compositions. Here we train a series of seven modern transformer models (GPT, GPT-2, GPT-Neo, GPT-J, BLMM, BART, and RoBERTa) for materials design using the expanded formulas of the ICSD, OQMD, and Materials Projects databases. Six different datasets with/out non-charge-neutral or EB samples are used to benchmark the generative design performances and uncover the biases of modern transformer models for the generative design of materials compositions. Our experiments show that the materials transformers based on causal LMs can generate chemically valid material compositions with as high as 97.61% to be charge neutral and 91.22% to be electronegativity balanced, which has more than six times higher enrichment compared to the baseline pseudo-random sampling algorithm. Our LMs also demonstrate high generation novelty and their potential in new materials discovery is proved by their capability to recover the leave-out materials. We also find that the properties of the generated compositions can be tailored by training the models with selected training sets such as high-bandgap samples. Our experiments also show that different models each have their own preference in terms of the properties of the generated samples and their running time complexity varies a lot. We have applied our materials transformers to discover a set of new materials as validated using density functional theory calculations. All our trained materials transformer models and code can be accessed freely at http://www.github.com/usccolumbia/MTransformer.
Export citation and abstract BibTeX RIS

Original content from this work may be used under the terms of the Creative Commons Attribution 4.0 license. Any further distribution of this work must maintain attribution to the author(s) and the title of the work, journal citation and DOI.
1. Introduction
Out of the almost infinite chemical design space of inorganic materials, there are only 262â242 experimentally synthesized crystal structures as deposited in the ICSD database [1] for now in April 2022. Intelligent computational algorithms are strongly needed to navigate the huge uncharted chemical space for discovering novel materials. Currently, there are three major new materials discovery strategies: the first one is experimental tinkering in which researchers manipulate a given composition, synthesize it, and characterize its structure or function [2]; the second approach uses computational models to generate new compositions, and uses crystal structure prediction algorithms to predict their structures, and then uses density functional theory (DFT) calculations to characterize their properties [3]; the third approach directly trains generative models for creating crystal structures for the downstream property prediction or simulation [4]. The first approach is too costly to explore the huge design space while the third approach is currently limited by the capability of existing crystal structure generation algorithms to generate stable structures. Considering the fact that most existing materials can be assigned to a limited number of prototypes, the emergence of template-based crystal structure prediction (TCSP) algorithms [5] has made it promising to explore new materials discovery using the composition generation and TCSP.
Here we propose to use the deep learning language models (LMs) for the generative materials composition discovery. Our work is inspired by the fact that a material composition or formula can be conveniently converted into a unique sequence of elements by assuming a specific element order (e.g. SrTiO3  Sr Ti O O O by the ascending element electronegativity) and that pretrained LMs have been widely used in the generation of natural language texts, molecules, and protein sequences. These pretrained self-supervised learning models such as BERT [6] and generative pre-trained transformer-3 (GPT-3) [7] are able to learn language/chemical grammars [8] for the text/molecule/protein generation [9, 10]. However, no such LMs have been used for the generation of inorganic materials.
Sr Ti O O O by the ascending element electronegativity) and that pretrained LMs have been widely used in the generation of natural language texts, molecules, and protein sequences. These pretrained self-supervised learning models such as BERT [6] and generative pre-trained transformer-3 (GPT-3) [7] are able to learn language/chemical grammars [8] for the text/molecule/protein generation [9, 10]. However, no such LMs have been used for the generation of inorganic materials.
There are several categories of pretrained LMs for the text generation as reviewed in [11] including masked LMs, causal LMs, prefix LMs, and encoder-decoder LMs. The masked LMs such as BERT are trained by predicting the masked tokens using the contextualized information, which is not directly in alignment with the text generation task. They however can be used in the encoder and decoder part for text generation models by exploiting their excellent bidirectional encoding capacities. Causal LMs such as GPT [12], GPT-2 [13], and GPT-3 are trained to calculate/predict the probability of the occurrence of several words given all preceding words, making them ideal for the text generation. However, they have the weakness of neglecting the bidirectional information. Prefix LMs such as UniLM [14] and XLNet [15] aim to combine the advantages of the bidirectional masked LMs and the unidirectional causal LMs in the text generation. A majority class of text generators such as T5 [16] and BART [17] belongs to the encoder-decoder LMs, which consist of stacks of both encoder and decoder layers. These models have all been used in the molecule or protein sequence generation but their performance for the inorganic composition generation is unknown.
Despite that crystal inorganic materials, organic molecules, and proteins are all composed of atoms, they have a distinct difference in terms of their building blocks and topology: organic molecules consist of atoms connected by bonds while protein sequences are composed of chains of amino acids. In contrast, crystal materials are periodic structures, of which each unit cell contains a repeating structural pattern. There is no strict chain of proteins or connected components of organic molecules. On the other hand, they can all be represented as sequences, and the models for the generative design of proteins and molecules can serve as the source of reference of developing generative models for the material composition generation.
Deep LMs have been used for the generation of molecules [18, 19]. Inspired by the GPT model, Bagal et al [18] trained a transformer-decoder on the task that predicts the next token using masked self-attention for the generation of druglike molecules. Rothchild et al [19] proposed their novel self-supervised pretraining method that enables transformers to make zero-shot select-and-replace edits, altering organic substances toward the desired property, which shows better performance and potential than graph-based methods. In [20], Kim et al combined a transformer encoder with a conditional variational autoencoder (cVAE) to achieve high-performance molecule generation. A similar generative VAE was also proposed in [21]. However, all these generative LMs do not explicitly model the generative process and work more like black-box generators. In addition to VAE-based models, researchers also use some Generative Adversarial Network (GAN)-based or Recurrent Neural Network (RNN)-based models to generate molecules. Guimaraes et al [22] proposed a method that combines GANs and reinforcement learning to achieve that while reinforcement learning biases the data generation process towards arbitrary metrics, the GAN component of the reward function ensures that the model still remembers information learned from data. De Cao and Kipf [23] adapted GANs to operate directly on graph-structured data and design experiments on the QM9 chemical database to validate the performance of their model.
LMs have also been applied for the protein sequence generation [24, 25]. Madani et al proposed an autoregressive transformer model named ProGen [24], an 1.2âbillion parameter conditional LM trained on a dataset of 280âmillion protein sequences. They also incorporated conditioning tags for taxonomic, functional, and locational information to enable the generation for targeted properties. Hesslow et al [26] trained decoder-only transformer models without any conditioning information for the protein sequence generation. Ingraham et al [27] proposed a conditional generative model for the protein sequence generation given 3D structures based on graph representations. More recently, Ram and Bepler developed the MSA-to-protein transformer, a generative model of protein sequences conditioned on protein families represented by multiple sequence alignments (MSAs). Compared with previous generative studies usually without rigorous performance evaluations, Ferruz et al [28] developed GPT-X based transformer models ProtGPT2 for generating de novo protein sequences. They found their generated protein sequences share some similarities with natural ones such as amino acid propensities and disorders. Linder et al [29] developed a LM that can maximize the fitness and diversity of synthetic DNA and protein sequences. Overall, except for the normalizing flow models, most generative models, such as autoregressive models (RNN/LSTM/Transformers), VAE, and GAN, have been applied for the protein generation [30]. However, compared to molecule deep generation model studies, there lacks standard benchmark datasets and performance evaluation metrics, which significantly hinder the development of generative protein sequence models.
Despite the success of deep LMs in the protein and molecule sequence generation, no studies have been reported successfully applied deep LMs to the inorganic materials composition generation except for our recent work on generative transformers [31]. Here, we develop and evaluate six materials transformers based on different LMs for the materials composition generation. We train all the models using the materials composition/formula data in the form of unlabeled expanded element symbol sequences from selected samples from ICSD/OQMD/Materials Projects (MPs) databases. Compared to natural language texts, inorganic materials composition sequences have strong constraints among the elements due to the requirements to form chemically valid and structurally stable periodic structures. This involves complex atomic interactions from ionic or covalent bonds and/or oxidation states of constituent elements. Effective materials composition generation models have to learn complex local and long-range dependencies and generation contexts, for which transformer neural network models excel at detecting and modeling. Our LM-based composition generators have an advantage over the heuristic or data mining element substitution models [32, 33] as they can consider the chemical context within the formulas rather than only element property compatibility. Our extensive generative composition design experiments show that the transformer-based materials generators can learn chemical grammars and achieve a high composition generation performance. Our additional experiments also show that materials transformers have different generation preferences or biases such as the tendency to generate materials compositions with  4 elements and with low numbers of atoms per element.
4 elements and with low numbers of atoms per element.
2. Materials and methods
2.1. Dataset
To evaluate the performance of our LM-based generators, we prepare six different datasets: Hybrid-mix, Hybrid-pure, Hybrid-strict, ICSD-mix, ICSD-pure, and Bandgap-30K.
The formulas in the Hybrid-mix dataset are selected from the ICSD/MP/OQMD databases with the number of elements less than 9, the number of atoms in the unit cell less than 100, and without fractional atom numbers for any element in the formula. While the Hybrid-mix dataset may contain a certain amount of materials that are not charge neutral or balanced electronegativity (EB), the Hybrid-pure dataset has samples selected from the Hybrid-mix dataset, which are charge neutral and have EB. The Hybrid-strict dataset is obtained using a similar method to the Hybrid-pure dataset, but we use the strict ICSD oxidation states of the elements to calculate the charge neutrality (CN) and EB, which is different (with more strict constraints) from the Hybrid-pure dataset and the dataset used in our previous study [31].
For the two ICSD datasets, the formulas in the ICSD-mix dataset are sampled from the ICSD database with the number of elements less than 9, the number of atoms in unit cell less than 100, and without fractional coordinates. It may contain formulas that do not meet the CN and EB criteria. Samples in the ICSD-pure dataset are selected from the ICSD-mix dataset, which satisfy CN and EB rules.
In addition, for the experiment of band gap prediction, we prepare a Bandgap-30K dataset, which contains 30â000 formulas from the MP database with band gaps above 1.98âeV. For those formulas with multiple phases, we include it if it has one phase with the band gap greater than 2.0âeV.
Overall, we get the Hybrid-mix, Hybrid-pure, Hybrid-strict, ICSD-mix, ICSD-pure, and Bandgap-30K dataset with total of 418â983, 257â138, 212â778, 52â317, 39â431, and 30â000 samples respectively. We divide all datasets into the training set, the test set, and the validation set in a ratio of 9/0.5/0.5, and the detailed number of each set is shown in table 1.
Table 1. Six datasets used in experiments. For Hybrid and ICSD datasets, -pure datasets only include selected samples with the neutral charge and balanced electronegativity; -mix datasets do not have such limits.
| Â | Hybrid datasets (ICSD + MP + OQMD) | ICSD datasets | MP dataset | |||
|---|---|---|---|---|---|---|
| Â | Hybrid-mix | Hybrid-pure | Hybrid-strict | ICSD-mix | ICSD-pure | Bandgap-30K |
| Total | 418â983 | 257â138 | 212â778 | 52â317 | 39â431 | 30â000 |
| Train | 398â033 | 244â281 | 202â139 | 50â755 | 37â459 | 28â500 |
| Validation | 10â475 | 6428 | 5319 | 1336 | 986 | 750 |
| Test | 10â475 | 6429 | 5320 | 1336 | 986 | 750 |
2.2. Pseudo-random composition generator
We build a pseudo-random composition generator as the baseline generation model. For all generated samples, we count the number of samples with different numbers of elements from 2 to 8. Then for each sample with the number of K elements, we generate the same number of composition samples with K elements. For these samples with K elements, We randomly pick the atom number from 1 to 20 for each of these K elements. This process ensures the distribution of binary, ternary, etc.
2.3. DFT calculations
We use the first-principles calculations based on the DFT using the Vienna ab initio simulation package to check the structural stability of the predicted materials [34â37]. The projected augmented wave pseudo-potentials with the plane-wave cutoff energy of 520âeV are used to treat the electron-ion interactions [38, 39]. The exchange-correlation functional is considered with the generalized gradient approximation based on the PerdewâBurkeâErnzerhof method [40, 41]. We set the energy convergence criterion as 10â5âeV, while the atomic positions are optimized with the force convergence criterion of 10â2âeVâÃ
â1. Furthermore, the Brillouin zone integration for the unit cells is computed using the Î-centered MonkhorstâPack k-meshes. The Formation energies (in eV/atom) of several materials are determined based on the expression in equation (1), where ![$E[\mathrm{Material}]$](https://content.cld.iop.org/journals/2632-2153/4/1/015001/revision2/mlstacadcdieqn3.gif) is the total energy per unit formula of the considered structure,
is the total energy per unit formula of the considered structure, ![$E[\textrm{A}_i]$](https://content.cld.iop.org/journals/2632-2153/4/1/015001/revision2/mlstacadcdieqn4.gif) is the energy of
is the energy of  element of the material, xi
indicates the number of Ai
atoms in a unit formula, and n is the total number of atoms in a unit formula(
element of the material, xi
indicates the number of Ai
atoms in a unit formula, and n is the total number of atoms in a unit formula( ),
),

2.4. Evaluation criteria
There are three main criteria we used in this paper, the validity, the uniqueness, the recovery rate and the novelty [3].
2.4.1. Validity
First, the two basic chemical rules of crystals, CN and EB percentages, are used to evaluate the validity of generated formulas. We use the method to calculate CN and EB proposed in [42] to obtain the percentages of generated samples that conform to these two rules. In addition, we check the stability of the generated samples to evaluate their validity performance using their predicted formation energies. The higher-energy region they are in, the lower quality they have.
2.4.2. Uniqueness
The uniqueness represents the ability of a generative model to generate unique formulas, which is used to calculate the percentage of the number of unique samples in the whole generated samples. The higher the uniqueness, the more diverse samples the model can produce.
2.4.3. Recovery rate and novelty
The recovery rate is used to estimate the percentage of generated formulas from the training set or the test set. The samples in the training set and test set are known, which means that the high recovery rate shows that this generative model has high performance on discovery materials. Another criterion related to the recovery rate is the novelty. The novelty of a generative model measures the percentage of the generated formulas that do not exist in the training or test sets.
2.5. Hyper-parameters tuning
Since the content and quantity of Hybrid datasets and ICSD datasets are very different, we tuned hyper-parameters for the Hybrid-mix dataset and the ICSD-pure dataset to choose appropriate hyper-parameters for Hybrid datasets and ICSD datasets respectively. Using the Hybrid-mix and the ICSD-pure datasets, we evaluated how the key hyper-parameters affect the generation performance on the MT-GPT2, the MT-BART, and the MT-RoBERTa models. We trained these models with different hyper-parameters, then generated about 10â000 formulas to evaluate their performance using two criteria, CN and EB. To find suitable parameters, We set default parameters for each model, then changed one of them each time to train models and evaluated their performance. The details of finetuning for all models can be found in supplementary file figures S2âS6.
2.6. Formation energy and band gap prediction models based on Roost
To evaluate the generator performances, we train two composition-based machine learning models. Both formation energy and band gap prediction use the dataset downloaded from the MP database [43]. The machine learning model we used is based on Roost, a graph message passing neural network as described in [44]. The training set of Roost-FE (formation energy) contains 125â613 unique compositions. For those compositions with multiple phases, we only keep the records with the lowest formation energies. The Roost-Bandgap model is trained with 113â501 samples. The formation energy roost model achieves an MAE of 70.181âeV while the band gap predictor achieves an MAE of 0.6645âeV as evaluated on the 10% hold-out test sets.
3. Results
3.1. Pretrain Transformer LMs for material composition generation
We select six different transformer-based LMs as implemented in the Huggingface package [45] to generate a series of material transformer (MT) generators. The group includes four GPT series LMs, BART [17], and RoBERTa [46]. In addition, we also use our previous work, BLMM [31], in our experiments to show the performance.
- GPT (Generative Pre-trained Transformer) model [12]: GPT is a transformer-based LM working on various Natural Language Processing (NLP) tasks with the unsupervised training. Original GPT uses 12-layer decoder-only transformer with masked self-attention, which is therefore powerful at predicting the next token in a sequence. Considering this property, we use GPT to generate our crystal formulas, and we call this GPT-based materials composition generation model as MT-GPT.
- GPT-2 model [13]: GPT-2 is a large transformer-based causal LM derived from GPT that is trained simply to predict the next word in large text corpses given all of the previous words within some text. Original GPT-2 models are trained with much more diverse text data with over an order of magnitude parameters than GPT. It uses a modified Byte Pair Encoding as input representation to combine the benefits of word-level LM with the generality of byte-level approaches. GPT-2 also has a few neural network architecture changes such as moving layer normalization to the input of each sub-block and adding a layer normalization to the final self-attention block. This model maps naturally to the crystal formula generation task. We call this GPT-2-based materials composition generation model as MT-GPT2.
- GPT-J model [47]: GPT-J is an open-source version of the multi-head GPT-3 model [7]. As an auto-regressive causal LM, it is originally used for the text generation task. GPT3 models use the same network architecture as GPT-2 except their use of alternating dense and locally banded sparse attention patterns in the transformer layers. Since the core ability of GPT-J is to take a string of the text and predict the next token, GPT-J is good at generating texts from a prompt. In this paper, We call the GPT-J-based materials composition generation model as MT-GPTJ.
- GPT-NEO model [48, 49]: GPT-Neo is an implementation of GPT3-like causal LM using the Mesh-TensorFlow library. The original architecture of GPT-Neo is similar to GPT-3 except that GPT-Neo uses local attention in every other layers with a window size of 256 tokens. GPT-Neo is trained as an auto-regressive LM, which means that it can also predict the next token as previous models. We call this GPT-Neo-based materials composition generation model as MT-GPTNeo.
- RoBERTa model [46]: RoBERTa is a dynamic masking pretrained LM based on the BERT model [6]. It achieves higher performance by applying an improved pretraining procedure over original BERT, which includes training with more epochs and larger mini-batches, removing the next sentence prediction objective, training on longer sequences and dynamically changing the masking pattern applied to the training data. We call this RoBERTa-based materials composition generation model as MT-RoBERTa.
- BART model [17]: BART combines the design of the transformer architecture with a bidirectional encoder (like BERT) and a left-to-right decoder (like GPT) to form a denoising autoencoder model. BART models are trained by corrupting text with a noising function and learning a model to reconstruct the original text. BART is effective on the text generation task while still performing well on the comprehension tasks. We call this BART-based materials composition generation model as MT-BART.
- BLMM [31] is a transformer-based generative LM for materials composition generation, which is based on the blank filling LM BLM [45]. It formulates the composition generation problem as a sequential probabilistic sequence rewriting problem, which allows it to directly model the generation process enabling its high interpretability and high efficiency in generation.
3.2. De novo generative design of materials composition
3.2.1. Model training and hyper-parameters
We prepare two sets of training datasets to train different MT models for the materials composition generation. The first set includes two datasets, the ICSD-mix dataset and the ICSD-pure dataset, which contain selected compositions from the ICSD database. The former includes samples that do not satisfy CN or EB while the latter contains only samples that satisfy both chemical criteria. To evaluate whether increasing the number of training samples can improve the generation performance, we also prepare the second set of datasets including Hybrid-mix, Hybrid-strict, and Hybrid-pure datasets with selected compositions from ICSD, MPs, and OQMD databases. The Hybrid-mix dataset includes all formulas from the three databases. The Hybrid-strict dataset are selected from the Hybrid-mix dataset, which contains only samples that satisfy CN and EB with the ICSD-oxidation assignments for all elements. The Hybrid-pure dataset has the same requirement except that the CN and EB are evaluated using more relaxed oxidation assignments for elements implemented as the default oxidation states in the SMACT package [42]. The detailed information of each dataset is in the section 2.1, and the detailed sample numbers of each dataset are shown in table 1.
For each dataset, we conduct hyper-parameter tuning to figure out the best hyper-parameters with reasonable tuning efforts (see section 2.5). We determine the training epochs based on the check of the learning curves of the training and validation loss to avoid overfitting. For each trained transformer model, we generate 50â000 candidate compositions that contain more than one and less than nine elements and the total number of atoms is smaller than or equal to thirty. We then check the percentages of these samples that satisfy CN and/or EB.
3.2.2. Generation of hypothetical material compositions
To evaluate whether our language MT models can learn the chemistry of inorganic materials (compositions) and use it to generate valid hypothetical formulas, we first compare the distributions of the generated samples by BLMM, MT-GPT2, and MATGAN [50] (a material composition generator) with respect to the training set. We represent each formula using the one-hot encoding as described in [3] and then map all the sample matrix representations into 2D space using the t-SNE algorithm. The results are shown in figure 1. As shown in figure 1(a), we find that the compositions of the ICSD materials in the training set are not evenly distributed with a shift towards the left half space. Figure 1(b) shows the distributions of the training set and samples generated by MT-GPT2 and BLMM, and the BLMM-generated samples have more overlaps with the training set compared to the MT-GPT2 generated ones. What is interesting is that both distributions of these two transformer generators are very different from the distribution of generated samples as distributed in figure 1(c) in which the training samples are organized into several clusters corresponding to materials families and the known materials (training and testing samples) are only a tiny portion of whole composition space and the MATGAN tends to generate very different samples.
Figure 1. The distributions of existing materials and hypothetical materials generated by BLMM, MT-GPT2, and MATGAN. The distributions are generated by calculating the one-hot representation for the compositions and then we use t-SNE to project them into 2-dimension space. (a) is the distribution of the training set. (b) is the distribution of the training set and generated samples by our BLMM and MT-GPT2. (c) is the distribution of the training set, the test set, and generated samples of MATGAN. Reproduced from [3]. CC BY 4.0.
Download figure:
Standard image High-resolution imageTo further illustrate the composition pattern of the materials generators, we combine the training set and the generated samples by BLMM, pseudo-random algorithm, MT-GPT, MT-GPT2, MT-GPTNeo, MT-GPTJ, MT-BART, and MT-RoBERTa and conduct the t-SNE dimension reduction to map the nine sets of samples into 2D space and then plot each set of compositions separately with the same coordinate system as shown in figure 2. First, we find that the BLMM-generated samples in (b) show the highest similarity to the training samples in (a) compared to all other generators. The next most similar distribution with regard to the training samples is from MT-BART, which has a similar rectangular shape but with fewer samples in the upper-left area. The GPT series transformers tend to generate compositions with similar distributions with variation in several local areas in the chemical space. For example, they all have sparse samples in the upper-left area, which may be due to they have difficulty generating compositions with fewer than five elements (see figure 2). The pseudo-random generator also shows a certain degree of distribution similarity with the training set because it uses its composition prototypes as the templates for composition generation, which means that it has the same number of binary, ternary, and quaternary samples as the training set. Finally, it is found that the samples generated by MT-RoBERTa have a very different distribution. This model is also the one that has the most difficulty to generate compositions with  30 atoms with
30 atoms with  8 elements.
8 elements.
Figure 2. The distributions of the training set and hypothetical materials generated by different materials transformers. The distributions are generated by calculating the one-hot representation for the compositions and then using t-SNE to project them into 2-dimension space. (a) Training samples; (b) BLMM; (c) random algorithm; (d) MT-GPT; (e) MT-GPT2; (f) MT-GPTNeo; (g) MT-GPTJ; (h) MT-BART; (i) MT-RoBERTa.
Download figure:
Standard image High-resolution image3.3. Evaluations of materials transformer generation performance using validity, uniqueness, recovery rate, and novelty
We evaluate the performance of our generative models for materials composition design and compare with the baseline random formula generator using four evaluation criteria including validity, uniqueness, recovery rate, and novelty as described in the section 2.4.
3.3.1. The CN and EB performance
Figure 3 shows the composition generation validity performance of seven transformer-based models compared them with the pseudo-random generator as evaluated on the ICSD-pure (37â459 samples with 95.94% CN and 93.12% CNâ+âEB) and ICSD-mix (50â755 samples with 78.40% CN and 72.36% CNâ+âEB) datasets. Note that all the percentages are calculated on the generated samples with less than or equal to 9 elements and 30 atoms. First, the CN and EB percentages of generated samples of all seven transformer models range from 65.87% to 97.54%, which are more than six times higher compared to the ones of the random generator (max CN: 10.13% and max CNâ+âEB: 5.91%). Out of the seven models, we find that overall, the MT-GPTNeo and MT-GPTJ have the best validity performance, which is followed by MT-GPT2, MT-BART, and MT-RoBERTa. The BLMM and MT-GPT models tend to have the lowest validity, even though their gaps with MT-BART and MT-RoBERTa are small. However, a close investigation of the filtering process shows that more than 45% of generated compositions have been filtered out for MT-BART, MT-RoBERTa, and the GPT series models while the BLMM filters out less than 0.3% samples, as shown in figure 9(a), MT models tend to generate formulas more than five elements, but we would like to filter these long formulas in the filtering process. Especially, we would like to note here that MT-RoBERTa's performance evaluated here is calculated on less than 10â000 generated samples as it has difficulty generating 50â000 candidate samples with atom numbers less than or equal to 30 and fewer than 9 elements within a reasonable amount of time (most samples are filtered out). The non-BLMM models tend to generate compositions with more than eight elements. In addition, these seven models show a big difference in terms of sample generation speed as shown in figure 8.
Figure 3. The comparison of the validity of materials composition generators on the ICSD-mix and ICSD-pure datasets. The percentages of CN and CNâ+âEB samples out of the training set and all generated samples by the generator models are used to represent validity.
Download figure:
Standard image High-resolution imageAnother performance trend we find is that the CN/EB validity percentages of samples generated by models trained on the ICSD-pure dataset are in general higher than those by the models trained on ICSD-mix except for the case of the CNâ+âEB percentages of MT-GPT2, MT-GPTJ, and MT-RoBERTa. After close examination, we find these exceptions are mainly due to the filtering process as discussed above.
We also compare the validity performance of our MT-GPTNeo model with our previously developed MATGAN models that are based on the generative adversarial network [3]. We find our MT-GPTNeo model achieves 97.54% CN and 84.64% EN (for the filtered samples with less than 31 atoms and 9 elements) compared to 80.3% CN and 70.3% EB achieved by the GAN model trained with ICSD-mix dataset, showing our models have about 14%â17% advantage. Similar performance advantages are also observed for our models trained with the ICSD-pure dataset. However, we want to note that this performance advantage is evaluated over the filtered samples.
To check whether increasing the training samples can improve the generator performance, we train our models on the three Hybrid datasets including Hybrid-mix, Hybrid-pure, and Hybrid-strict which have 398â033, 244â281, and 202â139 training samples respectively. The model hyper-parameters are also tuned with the new datasets. The final performances are shown in table 2. First, we find that the CN and EB percentage of the training set of Hybrid-mix is much lower than those of the ICSD-mix with only 61.67% CN and 50.61% respectively. However, the validity percentages of all models trained on the Hybrid-mix dataset are much higher than those of the training set. Especially GPT series models achieve CN percentages higher than 92% and EB percentages higher than 78%. These unexpected high validity percentages are due to the filtering process: they are good at generating chemically valid material compositions with less than 9 elements and 31 atoms while they also generate a large percentage of large formulas with many atoms ( 30) and elements (
30) and elements ( 8). Then, the Hybrid-pure dataset has higher CN (83.48%) and EB (75.94%) than the Hybrid-mix dataset. And it helps to improve the CN percentages of MT-GPTNeo, MT-GPTJ, MT-BART, and MT-RoBERTa and the EB percentages of MT-GPTNeo and MT-BART. Finally, the training set of the Hybrid-strict dataset has the highest CN (99.54%) and EB (99.25%). Almost all models except MT-GPTNeo achieve the highest CN and EB, indicating that high quality training samples contribute to the high validity performance of the trained models. Overall, we found that the MT-GPTJ has the best performance with CN of 96.68% and EB of 90.33% on Hybrid-mix, CN of 97.54% on Hybrid-pure, and CN of 97.61% on the Hybrid-strict dataset. We also found that MT-BART and MT-RoBERTa on the Hybrid-mix dataset have the lowest validity performance compared to those of GPT series.
8). Then, the Hybrid-pure dataset has higher CN (83.48%) and EB (75.94%) than the Hybrid-mix dataset. And it helps to improve the CN percentages of MT-GPTNeo, MT-GPTJ, MT-BART, and MT-RoBERTa and the EB percentages of MT-GPTNeo and MT-BART. Finally, the training set of the Hybrid-strict dataset has the highest CN (99.54%) and EB (99.25%). Almost all models except MT-GPTNeo achieve the highest CN and EB, indicating that high quality training samples contribute to the high validity performance of the trained models. Overall, we found that the MT-GPTJ has the best performance with CN of 96.68% and EB of 90.33% on Hybrid-mix, CN of 97.54% on Hybrid-pure, and CN of 97.61% on the Hybrid-strict dataset. We also found that MT-BART and MT-RoBERTa on the Hybrid-mix dataset have the lowest validity performance compared to those of GPT series.
Table 2. Comparison of generator performances of models trained with Hybrid datasets.
| Â | Hybrid-mix | Hybrid-pure | Hybrid-strict | |||
|---|---|---|---|---|---|---|
| Model | CN | CN + EB | CN | CNâ+âEB | CN | CNâ+âEB |
| TrainingSet | 61.67% | 50.61% | 83.48% | 75.94% | 99.54% | 99.25% |
| MT-GPT | 92.24% | 78.29% | 91.51% | 77.76% | 92.21% | 84.57% |
| MT-GPT2 | 92.96% | 79.79% | 87.82% | 70.83% | 96.99% | 92.81% |
| MT-GPTNeo | 93.84% | 84.37% | 97.29% | 91.40% | 94.69% | 77.39% |
| MT-GPTJ | 96.98% | 90.33% | 97.54% | 87.05% | 97.61% | 91.22% |
| MT-BART | 81.10% | 62.83% | 85.23% | 70.07% | 88.01% | 80.47% |
| MT-RoBERTa | 71.16% | 61.00% | 84.66% | 60.26% | 93.68% | 80.81% |
Note: The bold values represent the best performance of CN and CN + EB of different models on three different datasets, Hybrid-mix, Hybrid-pure, and Hybrid-strict.
As we mention earlier, the Hybrid datasets are datasets with samples ranging from 200â000 to 40â000, while the ICSD datasets are datasets with samples from 35â000 to 53â000. Therefore, we also compared the model performance trained by the Hybrid datasets and the ICSD datasets to investigate the effect of the quantity of samples on the generation performance. It is found that the CN and EB percentages of MT-GPT are not only increased from 81.01% and 66.54% to 92.24% and 78.29% on the Hybrid-mix, but also increased from 81.47% and 71.96% to 91.51% and 77.76% on Hybrid-pure. For MT-GPT2 and MT-RoBARTa, their CN and EB percentages have improved on the Hybrid-mix dataset compared to the ICSD-mix dataset. As for MT-GPTJ and MT-Bart, they achieve better CN and EB percentages on the Hybrid-pure dataset than those on the ICSD-pure dataset.
3.3.2. The stability performance
We also evaluate the materials composition generation validity performance by checking the stability of the generated compositions by all MT models and the pseudo-random generator as partially represented by their predicted formation energies. We train a Roost [44] based formation energy machine learning prediction model (see section 2.6) using all the MP samples. The formation energy distributions of the ICSD-pure training set, the generated samples of all MT models and random samples are shown in figure 4. First, we find that the formation energies of the training set are mostly less than zero eV and the shape of the distribution looks like a standard violin while the distribution of the random samples is very different from the training set with a large percentage of samples located in the near-zero eV area. Out of the six MT models and BLMM, the generated samples of MT-RoBERTa are of lower quality as they are more located in the high-energy region. While the samples of BLMM, MT-GPT and MT-GPT2 models show higher similarity in terms of the distribution shape to the training samples, the samples generated by MT-GPTJ, MT-GPTNeo, and MT-BART tend to have lower formation energy. The best quality of generated samples as represented by the predicted formation energy comes from the MT-GPTJ model, which has a peak density of samples around -15âeV for formation energy per atom. Overall, figure 4 shows that our MT models can generate chemical valid samples with negative formation energies.
Figure 4. The comparison of formation energy per atom distributions of the training samples, samples from the pseudo-random generator, and generated samples by different models trained on the ICSD-pure dataset.
Download figure:
Standard image High-resolution image3.3.3. The recovery rate performance
Another way to check the validity of generators is to check the number of generated samples that do not exist in the training set and exist as known materials in the leave-out dataset or third-party experimental or computational databases. The criterion is sometimes calculated as the recovery rate. We check the 56â162 generated samples of MT-GPTJ and find that 196 samples exist in one of the three databases, ICSD/OQMD/MP. In terms of holdout recovery rate, our model's performance (overall 0.81%) is much lower compared to those of the BLMM model, which achieves 100% for binary materials, 63.37% for ternary materials, and 29.17% for quaternary ones. It is also lower than the MATGAN model which has binary, ternary, and quaternary compounds recovery rates of only 82.7%, 31.2%, and 5.2%. The reason is that our transformer-based LMs tend to generate compositions with more than five elements (see figure 9(b)).
3.3.4. The uniqueness performance
Another important performance measure of generative models is the uniqueness, which represents the percentage of unique samples out of all generated samples [51]. Here for five MT-GPTNeo models trained on the five datasets, we calculate the uniqueness percentages for every 200 generated samples up to 10â800 samples with  30 atoms and
30 atoms and  8 elements. The results are shown in figure 5. First, we find that all five MT-GPTNeo models show high uniqueness: after generating 10â800 filtered samples, the uniqueness percentages remain around or above 50%: ICSD-mix (59.91%), ICSD-pure (61.38%), Hybrid-mix (55.06%), Hybrid-strict (47.69%), and Hybrid-pure (28.27%). Another interesting observation is that the models trained on the ICSD-mix and ICSD-pure datasets dominate in terms of the uniqueness, which is probably due to the fact that these two datasets have much fewer training samples (50â755 and 37â459 versus more than 200â000 for hybrid datasets). The more training samples, the more strict language constraints the models learn, and thus the lower the diversity/uniqueness in the generated samples. Among all the three Hybrid models, the model trained with the Hybrid-mix has the highest uniqueness since it contains diverse samples that do not satisfy CN/BN chemical rules. The uniqueness difference of these three models can be attributed to their different distributions among the training sets.
8 elements. The results are shown in figure 5. First, we find that all five MT-GPTNeo models show high uniqueness: after generating 10â800 filtered samples, the uniqueness percentages remain around or above 50%: ICSD-mix (59.91%), ICSD-pure (61.38%), Hybrid-mix (55.06%), Hybrid-strict (47.69%), and Hybrid-pure (28.27%). Another interesting observation is that the models trained on the ICSD-mix and ICSD-pure datasets dominate in terms of the uniqueness, which is probably due to the fact that these two datasets have much fewer training samples (50â755 and 37â459 versus more than 200â000 for hybrid datasets). The more training samples, the more strict language constraints the models learn, and thus the lower the diversity/uniqueness in the generated samples. Among all the three Hybrid models, the model trained with the Hybrid-mix has the highest uniqueness since it contains diverse samples that do not satisfy CN/BN chemical rules. The uniqueness difference of these three models can be attributed to their different distributions among the training sets.
Figure 5. The uniqueness of the MT-GPTNeo model trained on ICSD-pure dataset.
Download figure:
Standard image High-resolution image3.3.5. The novelty performance
We also check the novelty of the composition generators, which calculates the percentages of samples that do not exist in the training set. All our six models achieve more than 97% overall novelty when trained over the ICSD-pure dataset. In contrast, the BLMM model achieves 97.66%, 96.11%, and 95.55% novelty for binary, ternary and quaternary compounds respectively, indicating comparable or slightly better capability to generate new materials.
To understand why some transformer models are better than the others, we compile the numbers of parameters, unique features of the neural network models, and their relative performances for six models in the supplementary file table S1. First, we find that the MT-RoBERTa and MT-BART models have the lowest performance (see table 2) due to the fact that both are masked LMs, which are trained to predict masked tokens instead of next tokens. Next, it is found that the MT-GPT and MT-GPT2 have much better performance than the previous two because these are causal LMs trained to predict the next token. This naturally facilitates the models' ability to generate valid token sequences by learning the dependency relationships among the tokens. Finally, we find the MT-GPTJ and MT-GPTNeo models achieve the best performance with significantly more parameters and a few network architectural innovations. Overall, the MT-GPTJ achieves the highest performance.
3.4. Conditional generative design of materials compositions with high bandgap
Conditional generation capability is highly desirable for function-oriented computational materials design. Flam-Shepherd et al [52] used this evaluation method for benchmarking molecule generator models. Therefore, to evaluate whether our LM can capture the composition rules for assembling high-bandgap materials to directed high-bandgap materials generation, we collect 30â000 formulas with band gaps above 1.98âeV from the MPs database (for those formulas with multiple phases, we include it if it has one phase with band gap greater than 2.0âeV). We call this the Bandgap-30K dataset. We train an MT-GPT2 composition generator and use it to generate 100â000 formulas from which 87â233 compositions satisfy the CN and EB requirements. We then use the composition-based band gap prediction model (see section 2.6) to predict the band gaps of these filtered hypothetical material compositions and plot their distribution against the band gap distributions of the training set and the whole MP samples. As shown in figure 6, the band gap distribution of our hypothetical materials is much closer to the high-bandgap training set compared to the band gap distribution of all MP samples, which indicates that the MT-GPT2 bandgap model has learned the implicit rules to generate high-bandgap materials.
Figure 6. Band gap distribution for (1) the whole Materials Projects (MPs) materials, (2) the training set of high-bandgap MP materials for the MT-GPT2 model; (3) the generated samples from the MT-GPT2 models trained on the MP dataset. The band gap distribution of the generated ones is much closer to the training set than the whole MP dataset, they tend to have higher band gaps.
Download figure:
Standard image High-resolution image3.5. New materials predicted by MT-GPT2 and validated using DFT
We use the Bandgap-30K dataset to train a MT-GPT2 model and use it to generate more than 100â000 material compositions. Then we predict their formation energy using the composition based formation energy prediction model (see section 2.6). When we get the formation energies, we calculate their total energy and predict their e-above-hull energies to rank these candidates. We then pick the top 100 formulas with the lowest predicted e-above-hull energy and apply our previous TCSP, a template based crystal structure prediction algorithm [5], to obtain the structures. For the predicted structures with the best quality scores, we run DFT relaxation to get the final structures and to calculate their formation energy and e-above-hull energy (see section 2.3). Table 3 shows the top 20 discovered new materials along with their formation energies. Out of the predicted structures, we identify two new crystal materials with the e-above-hull energy of 0âeV, and their structures are shown in figure 7.
Figure 7. Candidate new structures discovered by our MT-GPT2 model with zero e-above-hull.
Download figure:
Standard image High-resolution imageFigure 8. Time for generating 1000 compositions by different models. The fastest model is BLMM while the slowest one is MT-RoBERTa.
Download figure:
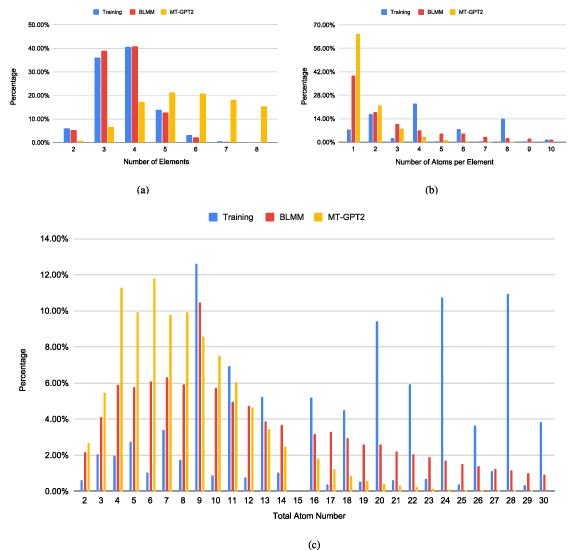
Standard image High-resolution imageFigure 9. The comparison of the generation preferences of training samples, BLMM, and MT-GPT2 in terms of generated composition properties. (a) Distributions of the element numbers in each composition; (b) distributions of the atom numbers for each element in all compositions. For the training set, the elements with more than 28 atoms are not counted in this plot. (c) Distribution of total atom number for each composition. For the training set, the compositions with more than 30 atoms are not plotted in this figure.
Download figure:
Standard image High-resolution imageTable 3. Twenty materials found with negative formation energy ( ) using DFT.
) using DFT.
| Formula |

| Formula |

|
|---|---|---|---|
| SrAlClO2 | â3.0077 | BaSrTiO3 | â2.5268 |
| LiMgBrF2 | â2.9039 | LiScNiF3 | â2.5214 |
| BaScSeF2 | â2.8662 | ScBeOF | â2.5074 |
| AlBrF2 | â2.8367 | KSrAlO3 | â2.5024 |
| LiMg2IF4 | â2.8118 | AlFeOF3 | â2.5015 |
| KSrScO3 | â2.7351 | Sc2AlZnO4 | â2.4719 |
| BaScO2 | â2.6801 | LiBeOF | â2.4639 |
| KBaScO3 | â2.6221 | SrBeSeF2 | â2.4584 |
| KBaAlO3 | â2.5924 | MgVHF4 | â2.4521 |
| RbBeOF | â2.5392 | KSrZrO3 | â2.4300 |
4. Discussion
We have applied a series of transformer-based pretrained LMs for the generative design of inorganic materials compositions. Our extensive experiments over five datasets with diverse sample sizes and quality (in terms of CN and EB) show that these materials transformer models are competent in generating chemically valid compositions or formulas while most of them have their own generation preference or bias. We find that MT-GPTJ overall has the best generation performance (after simple filtering) in terms of validity and generation speed.
We check the time complexity of the generators as shown in figure 8. We count the amount of time (in seconds) that each model needs to generate 1000 compositions without any filtering. It is found that the BLMM model has the fastest generation with 145âs. The second fastest models are MT-GPT2, MT-GPTJ and MT-GPTNeo which use 219, 449, 366âs respectively. The slowest generators include MT-GPT, MT-BART, and MT-RoBERTa which are almost 7.6 to 13.6 times slower compared to BLMM. We also find that the generator models vary a lot in terms of generating qualified candidates that have  30 atoms and
30 atoms and  8 elements. Figure S8 in the supplementary file shows the number of qualified candidate compositions generated by different models within 25â000 loops, each of which a sequence of 256 tokens is generated and then partitioned into multiple compositions. We find that MT-RoBERTa has an extremely low yield rate in generating qualified compositions compared to other transformer models. It is surprising that the MT-GPT model trained on the ICSD-pure dataset also has the difficulty to generate such qualified individuals. Both of them tend to generate formulas with
8 elements. Figure S8 in the supplementary file shows the number of qualified candidate compositions generated by different models within 25â000 loops, each of which a sequence of 256 tokens is generated and then partitioned into multiple compositions. We find that MT-RoBERTa has an extremely low yield rate in generating qualified compositions compared to other transformer models. It is surprising that the MT-GPT model trained on the ICSD-pure dataset also has the difficulty to generate such qualified individuals. Both of them tend to generate formulas with  8 elements.
8 elements.
Another potential factor that affects the generator performance is the training set size. To check this issue, we train 15 MT-GPT2 models with training set sizes ranging from 1000 to 377â084. Their generation performances in terms of validity as represented by the CN and EB percentages are shown in the supplementary file figure S7. A general trend we find is that increasing the training set size can lead to better generator performance. Another major decision in materials transformer training is to determine the optimal training epochs for each model over different datasets so that overfitting can be avoided. In the supplementary file, figure S7(b) shows the learning curves of training and validation errors for training MT-GPT2. It is found that after around 700 epochs, the training process starts to overfit as the validation loss begins to dominate the training loss. However, we observe that the corresponding CN/EB percentages do not degrade much despite the occurrence of the overfitting.
To further examine the generation capabilities and potential bias of different materials transformers, we train the BLMM and MT-GPT2 models using the Bandgap-30K dataset as defined in section 3.4 (Conditional generation) and generate 89â321 and 87â232 samples respectively. We then plot the distributions of the number of elements within compositions, the number of atoms per element, and the total number of atoms within a composition as shown in figure 9. First, we find that the distribution of the number of elements within the training compositions are highly imbalanced (figure 9(a)) with the highest percentage of ternary and quaternary materials followed by quinary and binary materials. There are very few samples with more than six elements. It is then interesting to find that the element number distribution of the BLMM-generated samples follows closely to the training set, indicating that the BLMM model tends to learn well of the chemical patterns from the training set. On the other hand, the MT-GPT2-generated samples have a very different distribution in terms of the element number of compositions: it tends to generate a large proportion of samples with more than four elements.
Next, figure 9(b) shows the distribution of the number of atoms per element within the samples. The training samples have a variety of atom numbers per element ranging from 1 to more than 10, with relatively small percentages of ones. In contrast, the samples generated by BLMM contain much more compositions with 1-atom elements. However, the MT-GPT2 model is even more biased as it tends to generate most of single-atom elements in the compositions (65%) followed by 2-atom elements (16%) (check yellow bars). It has a low probability to generate samples with more than four-atom elements. We find the generation preferences of MT-GPT2 also apply to all other transformer-based models except BLMM.
We further check the distribution of the total atoms within a composition as shown in figure 9(c). First, it is found that the training set has many samples with a large number of atoms with peaks at 9, 20, 24, and 28. In contrast, the total atom numbers of the samples generated by GPT2 peak at 4, 5, 6, 7, 8, and 9, which are much smaller than the training set. It can barely generate samples with total atom numbers more than 20. On the other hand, the BLMM model has a more balanced generation capability: while it also peaks at 9, 4, 5, 6, 7, 8, 10, and 11 in terms of the total atom number within compositions, it can still generate large percentages of samples with more than 20 atoms. It is also interesting to notice that there are no materials with 15 atoms within the formula.
5. Conclusion
We develop and benchmark seven modern deep transformer LMs for crystal material composition generation. Our extensive experiments show that the transformer-based LMs can generate chemically valid hypothetical material compositions as shown by the high percentages of generated charge neutral and EB samples. However, it is not clear whether these compositions can be synthesized into stable structures. This especially may become a concern when a generative model tends to generate compositions with a large number of elements. While several machine learning models have been developed for synthesizability prediction [53], formation energy prediction [54], and e-above-hull calculation, these models and algorithms usually require the availability of the crystal structures which are not available for composition generators that we propose here. To do that, we can use the recently developed TCSP algorithms [5, 55] or the deep learning-based [56], and global optimization-based crystal structure prediction tools [57, 58] to predict the crystal structures for the generated hypothetical compositions by our materials transformer models. Together, we are able to explore and discover new materials in a much larger area of the almost infinite chemical design space.
Data availability statement
The data that support the findings of this study are openly available at the following URL/DOI: http://github.com/usccolumbia/mtransformer.
The raw materials composition dataset is downloaded from MaterialsProject.org, OQMD.org and ICSD. The training/validation/testing datasets used in our model training are deposited at figshare.com at this url https://figshare.com/articles/dataset/MT_dataset/20122796.
Code availability
The materials transformer language models are trained using the Huggingface transformer package. The trained models and the training and generation code can be downloaded at http://github.com/usccolumbia/mtransformer.
Funding
The research reported in this work was supported in part by National Science Foundation under the Grant Nos. 1940099, 1905775 and 2110033. The views, perspectives, and content do not necessarily represent the official views of the NSF.
Author contributions
Conceptualization, J H; methodology, J H, N F, L W, Q L, D S, Y S; software, N F, J H, L W, Y S, Q L; resources, J H; writingâoriginal draft preparation, J H, F N, L W, D S, R X, S S O; writingâreview and editing, J H, F N, L W, D S; visualization, N F, J H, L W, Y S, Q L, D S; supervision, J H; funding acquisition, J H.
Conflict of interest
The authors declare that they have no competing interests.
Supplementary data (0.5 MB PDF)