ISSN: 1838-7640Theranostics
Theranostics 2019; 9(4):1085-1095. doi:10.7150/thno.29719 This issue Cite
Research Paper
Zwitterionic near-infrared fluorophore-conjugated epidermal growth factor for fast, real-time, and target-cell-specific cancer imaging
1. Research Institute, National Cancer Center, 323 Ilsan-ro, Goyang, Gyeonggi 10408, Republic of Korea
2. Gordon Center for Medical Imaging, Department of Radiology, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Abstract
Epidermal growth factor receptor (EGFR) is overexpressed in many types of cancers, which is associated with metastatic potential and poor prognosis in cancer patients. Therefore, development of EGFR-targeted sensitive imaging probes has been a challenge in tumor targeting, image-guided cancer surgery, patient-selective anti-EGFR therapy, and efficient targeted therapies.
Methods: We synthesized a zwitterionic near-infrared fluorophore (ATTO655)-conjugated epidermal growth factor (EGF) as a novel activatable molecular probe. Fluorescence OFF/ON property and EGFR-targeting specificity of EGF-ATTO655 as well as its utility in real-time near-infrared (NIR) fluorescence imaging of EGFR-positive cancers were evaluated using in vitro and in vivo studies.
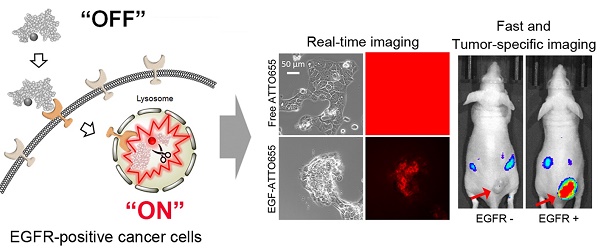
Results: When conjugated to EGF, the fluorescence of ATTO655 quenched efficiently by photo-induced electron transfer (PET) mechanism between the conjugated dyes and nearby amino acid quenchers (tryptophan/tyrosine residues), which was stably maintained at physiological pH and in the presence of serum for at least 17 h. The fluorescence of EGF-ATTO655 turned on by receptor-mediated endocytosis and subsequent disintegration of EGF in EGFR-positive A431 cancer cells, thereby enabling specific and real-time fluorescence imaging of EGFR-positive cancer cells. Consequently, EGFR-positive tumors could be clearly visualized 3 h post-injection with a significantly high tumor-to-background ratio (TBR = 6.37).
Conclusion: This PET mechanism-based OFF/ON type of EGF probe showed great potential for rapid, real-time, and target-cell-specific imaging of EGFR-overexpressing cancers in vitro and in vivo.
Keywords: Epidermal growth factor, photo-induced electron transfer, tumor-specific targeting, real-time cancer imaging
Introduction
Epidermal growth factor receptor (EGFR) is a transmembrane protein of the tyrosine kinase receptor family and plays an important oncogenic role in the growth, survival, migration, and invasion of cancer [1-3]. EGFR is overexpressed on the surface of various cancers including skin, breast, ovary, bladder, prostate, head and neck, non-small cell lung cancers, and is correlated with metastatic potential and poor prognosis in cancer patients. EGFR is, therefore, an important target in cancer patients for monitoring prognosis, targeted chemotherapy, monoclonal antibody therapy, and immune therapy [4-8].
Many efforts have recently been made to conjugate near-infrared (NIR) fluorophores on anti-EGFR antibodies, and cetuximab labeled with IRDye800CW is currently in clinical trials for image-guided surgery of EGFR-positive cancers [9-11]. Despite of potential utility in fluorescence-guided tumor resection [12-15], antibody conjugates have intrinsic limitations in in vivo cancer imaging due to the large size (MW ~ 150 kDa), resulting in slow clearance from the body and prolonged generation of background fluorescence as well as poor tissue permeability that lower the tumor-to-background ratio (TBR) after systemic administration [15]. Moreover, real-time fluorescence identification of EGFR-positive cancer cells is limited because cetuximab-NIR fluorophores are always on (i.e., fluorescent) regardless of binding to the target cells, and thereby it is impossible to discriminate fluorescence signals of the target from the off-target (i.e., non-specific uptake) [15]. Therefore, the time point for tumor imaging should be optimized depending on the clearance rate of each agent to reduce background signals [16]. This is a general problem even for small sized EGF [16-19]. EGF shows high binding affinity and relatively good tissue permeability owing to its small size, however, for example, the TBR values obtained in HCT116 xenograft tumor mice using small-sized EGF-quantum dot conjugates were less than 3 during 24 h post-intravenous injection [16].
To overcome this limitation, herein, we introduce a zwitterionic NIR fluorophore-conjugated EGF as a new activatable probe for fast and target-cell-specific imaging of EGFR-positive cancer. EGF is consisted of 53-amino acids including two tryptophan (Trp) and five tyrosine (Tyr) residues [20]. We hypothesize that when a zwitterionic dye is conjugated to EGF, its fluorescence could be efficiently quenched by the photo-induced electron transfer (PET) mechanism between the dye and nearby amino acid quenchers (Trp/Tyr residues) [21-22]. Given that the distance between a fluorophore and quenchers is within 1 nm scale [23], the quenching efficacy is highly sensitive to the small changes in the tumor microenvironment, where the fluorescence should selectively turn on by receptor-mediated endocytosis and subsequent disintegration of EGF in EGFR-positive cancer cells, thereby enabling specific and real-time fluorescence imaging of cancer cells.
Materials and Methods
Materials
Human epidermal growth factor (EGF), Amicon ultra centrifugal filter unit (MWCO 3 K), sodium dodecyl sulfate (SDS), and 2-mecaptoethanol (ME), dithiothreitol (DTT) were obtained from Sigma-Aldrich (St. Louis, MO, USA). ATTO655-COOH (λex/λem = 663/684 nm) and ATTO655-N-hydroxysuccinimidyl ester (ATTO655-NHS ester) were obtained from ATTO TEC (Siegen, Germany). Lysotracker (Blue DND-22), fetal bovine serum (FBS), Lab-Tek II chambered coverglass, DMEM, and RPMI 1640 were purchased from Thermo Fisher Scientific Inc. (Waltham, MA, USA). PD-mini Trap G25 column was purchased from GE Healthcare (Little Chalfont, UK). 4′,6-diamidino-2-phenylindole (DAPI) was obtained from Vector Laboratories Inc. (Burlingame, CA, USA).
Synthesis of EGF-ATTO655 conjugates
Human EGF (0.5 mg, 8 μmol) and ATTO655-NHS ester (16 μmol) were dissolved in phosphate-buffered saline (PBS; pH 7.4, 10 mM, NaCl, 137 mM; 0.3 mL) and reacted for 1 h at 25 ºC. The reaction mixture passed through a PD-mini Trap G25 column to remove byproducts and unbound dyes. Next, purified solution of EGF-ATTO655 conjugates was concentrated using Amicon Ultra centrifugal filters and then stored at 4 °C before use.
Characterization of EGF-ATTO655 conjugates
To analyze degree of labeling (i.e., number of dyes per an EGF), EGF-ATTO655 was diluted with PBS containing 1% SDS and 1 mM ME (final concentration: 5 μM dye equivalent.) for denaturation of the conjugate, and its absorbance at 663 nm was measured and compared with the standard curve of free ATTO655-COOH. EGF concentration in the solution was calculated using the molar extinction coefficient of EGF (i.e., 18825 M-1cm-1 at 280 nm) which was based on peptide sequence from Expasy (ProtParam tool).
To check fluorescence dequenching upon denaturation of EGF-ATTO655, a stock solution of EGF-ATTO655 was diluted with PBS or PBS containing denaturing reagents (1% SDS + 1 mM ME) to the final concentration of 5 μM dye equivalent. Then, their absorption and fluorescence spectra were analyzed. As a control, free dye (ATTO655-COOH) was dissolved in PBS containing 1% SDS + 1 mM ME at 5 μM, and its absorption and fluorescence spectra were measured and compared with that of the conjugate.
NIR fluorescence imaging of EGF-ATTO655 in PBS, EGF-ATTO655 in PBS containing 1% SDS + 1 mM ME, and free ATTO655 in PBS containing 1% SDS + 1 mM ME (final concentration: 5 μM dye equivalent.) were performed using a IVIS Lumina XR Imaging System (PerkinElmer; λex 620/20 nm, λem 670/40 nm) to visualize the quenched and dequenched status of the conjugate.
Analysis of dye-conjugated site in EGF-ATTO655
For peptide mapping, EGF and EGF-ATTO655 were precipitated with cold acetone and their disulfide bonds were reduced by 10 mM DTT. And both of them were alkylated with idoacetamide (IAA) and digested with Glu-C endoprotease in 25 mM NH4CO3 for 12 h at 37 ºC. Digested fragments were prepared for MS analysis after cleaned up using C18 ZipTip (Millipore) and then evaporated with vacuum concentrator. The isolated peptides were analyzed by a Q ExactiveTM hybrid quadrupole-orbitrap mass spectrometer equipped with an Ultimate 3000 RSLCnano system (Thermo Fisher Scientific). Raw data files of EGF and EGF-ATTO655 obtained from LC-MS/MS analysis was matched with Swissprot-Human database using Proteome Discoverer 2.1 software and identified parameters with fixed modification for carbamidomethyl cysteine (+57.021 Da / C) and variable modifications for ATTO655 conjugated lysine (+509.198 Da / K). The false discovery rate (FDR) of peptide identification is less than 1 % on the peptide level and filtered with the high peptide confidence. Detailed information on the analytic methods is described in the supporting information.
Stability of EGF-ATTO655 in serum condition
To test stability of the quenched state of EGF-ATTO655, a stock solution of the conjugate was diluted in PBS (pH 7.4) and PBS (pH 7.4) containing 10% FBS at a final concentration of 1 μM dye equiv., respectively. As a control, free ATTO655-COOH dye at the same concentration was also diluted in PBS. Fluorescence intensities of the sample solutions (λex 600 nm, λem 684 nm) were measured every 1 h for 17 h using a multifunctional microplate reader.
Fluorescence turn-on of EGF-ATTO655 upon enzymatic degradation
EGF-ATTO655 (1 μM dye equivalent) in phosphate buffer (pH 5.0) was treated with either 20 μg/mL proteinase K or 5 mM DTT, and then its change in fluorescence intensities with time was measured every 30 min for 18 h (λex 600 nm, λem 684 nm). Fluorescence intensities of EGF-ATTO655 in phosphate buffer (pH 5.0) without the enzyme and DTT were also measured for 18 h as a control.
Evaluation of EGFR-targeting specificity of EGF-ATTO655
The A431 (human epidermoid carcinoma; EGFR-positive) and NCI-H460 (human non-small-cell lung carcinoma; EGFR-negative) cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA), and maintained in DMEM and RPMI-1640 cell culture media supplemented with 10% FBS, respectively [24, 25].
These cells were seeded on a 4-well Lab-Tek II chambered coverglass at 50,000 cells per well and incubated overnight. Free ATTO655 dye and EGF-ATTO655 were diluted with cell culture medium to obtain 1 μM dye equivalent. A431 and NCI-H460 cells were treated with free ATTO655 dye or EGF-ATTO655 for 5 h. For competition assay of receptor binding, A431 cells were pre-incubated with excess amount of unlabeled EGF (100 μg) for 30 min and then treated with EGF-ATTO655 (1 μM dye equivalent). After washing the cells three times with PBS solution, lysosomes of the cells were stained with Lysotracker. Then, confocal fluorescence imaging of the cells (Lysotracker: λex 405 nm, λem 410-585 nm, ATTO655: λex 633 nm, λem 638-759 nm) were performed using a confocal laser scanning microscope (Carl Zeiss LSM 780 META, Jena, Germany).
Real-time NIR fluorescence imaging of EGFR-positive cancer cells
EGFR-positive A431 cells were seeded on an 8-well Lab-Tek II chambered coverglass at 20,000 cells per well for real-time fluorescence imaging study. A431 cells were treated with free ATTO655 dye or EGF-ATTO655 at the concentration of 1 μM dye equivalent. NIR fluorescence images (λex 640/30 nm, λem 690/50 nm) of the cells were obtained every 15 min without washing steps for 6 h using a Live Cell Imaging System (Axio observer Z1, LD Plan-Neofluar 20x/0.4 Korr Ph2 M27, Carl Zeiss, Jena, Germany). After completion of the live cell imaging, the cells were washed three times with cell culture medium to remove the free dye and the conjugates remaining in the extracellular space, and then fluorescence images of the cells were acquired once more. All images were acquired using the same microscope settings in order to ensure reproducibility.
In vivo and ex vivo NIR fluorescence imaging
NCI-H460 and A431 cells (5 × 106 cells/200 μL) were subcutaneously implanted into the right hind flank of each mouse (Balb/c-nu, female). When tumor sizes reached ~190 mm3, the mice were involved in in vivo NIR fluorescence imaging study. The mice bearing EGFR-negative NCI-H460 tumors received intravenous injections of the EGF-ATTO655 (n = 3, 100 µg/100 µL PBS/mouse) or PBS (n = 3, 100 µL PBS/mouse) via tail vein. Also, the mice bearing EGFR-positive A431 tumors received intravenous injections of the EGF-ATTO655 (n = 3, 100 µg/100 µL PBS/mouse) or PBS (n = 3, 100 µL PBS/mouse). For competition assay of receptor binding, three mice bearing A431 tumors received intravenous injection of 100 µL PBS solution containing both unlabeled EGF (300 μg) and EGF-ATTO655 (100 μg). Then, NIR fluorescence imaging of the mice (λex = 620/20 nm, λem = 670/40 nm) were carried out using the IVIS Lumina XR imaging system at 3 and 24 h post-injection. For biodistribution analysis, the mice were sacrificed at 24 h post-injection and ex vivo NIR fluorescence imaging of the tumors and organs (kidneys, spleen, and the liver) were carried out. After that, the collected tumor tissues were immediately frozen and cryo-sectioned at 7 μm thickness. The nuclei of the cells in the tumor sections were counter-stained with DAPI, and then fluorescence images of the tumor sections (λex 633 nm and λem 638-758 nm for EGF-ATTO655; λex = 405 nm and λem 410-482 nm for DAPI) were observed using a confocal scanning laser microscope.
All the animal studies were approved by the Institutional Animal Care and Use Committee of National Cancer Center.
Statistical Analysis
Data are expressed as mean (standard deviation). Student's t-test was used to determine significant difference between test groups.
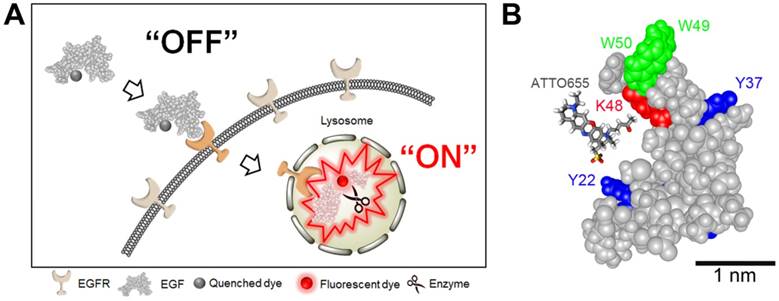
(A) Schematic illustration of tumor-specific fluorescence turn-on of quenched EGF-ATTO655 conjugate in EGFR-overexpressing cancer cells. Quenched status of EGF-ATTO655 is stably maintained in the extracelluar space by PET interaction between the conjugated ATTO655 and amino acid quenchers (Trp and Tyr), but its NIR fluorescence is turned on after receptor-mediated endocytosis and subsequent disintegration of 3-dimentional structure of EGF-ATTO655, thereby enabling target-cell-specific and real-time fluorescence imaging of EGFR-positive cancer cells. (B) 3D drawing of EGF-ATTO655. Zwitterionic NIR fluorophore, ATTO655 was conjugated to Lys48 (red, K48) which is located in close proximity with Trp (green, W49, W50) and Tyr (blue, Y22, Y37) residues in EGF.
Results and Discussion
Synthesis and characterization of EGF-ATTO655
As shown in Figure 1, ATTO655 was introduced to the Lys of EGF via conventional NHS chemistry. ATTO655-NHS ester was reacted with the amine group of the Lys residues in EGF at the reaction ratio of 2:1. According to the analysis of UV/Vis absorbance of EGF-ATTO655, degree of labeling (i.e., the number of the conjugated dyes per an EGF) was calculated to be 0.78. The molecular weight of EGF-ATTO655 measured by MALDI-TOF was 6,888.958 Da, (Figure S1), supporting that the number of the conjugated dye per an EGF peptide was mostly one, instead of two dyes. Between the two lysine residues (K28, K48) in EGF, K48 was analyzed as the main conjugation site of the dye (Figure 1B, Table S1 and S2). According to the structure of EGF, the K48 residue is located in the highly flexible C-terminal region, whereas the other lysine residue, K28, is located in the structurally rigid EGF core (i.e., the antiparallel β-sheet of EGF with high structural rigidity). The high flexibility of the K48 residue may be the reason why the ATTO655 dye was mainly conjugated to this site instead of K28. The three-dimensional structural prediction for EGF-ATTO655/EGFR complex (Figure S2) showed that the interaction between EGFR and the EGF core is retained after ATTO655 conjugation, and the ATTO655-conjugated C-terminal region with high structural plasticity does not seem to interrupt either the binding of EGF to EGFR or the structure of EGFR.
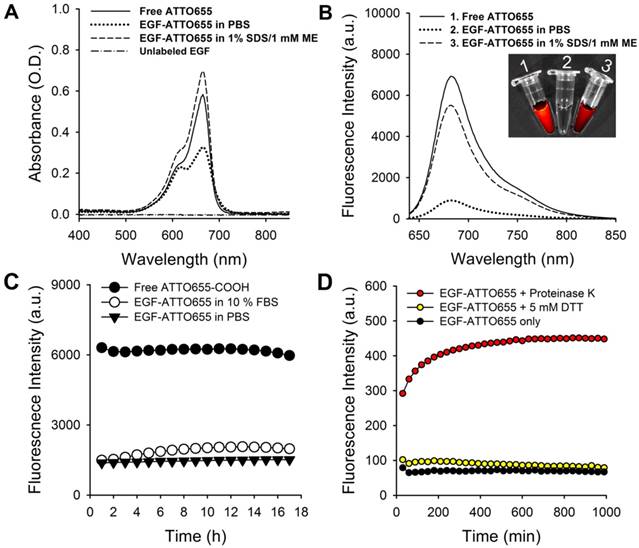
EGF-ATTO655 in PBS (pH 7.4) showed broadened and lowered UV/Vis absorption spectrum than that of the free dye (Figure 2A). UV/Vis absorption spectrum of EGF-ATTO655 becomes similar to that of free dye when 3-dimensional structure of the conjugate was disintegrated with PBS containing anionic surfactant (1% SDS) and reducing agent (1 mM ME). Likewise, fluorescence intensity of EGF-ATTO655 in PBS (pH 7.4) was 7.8-fold lower than that of free ATTO655 dye at equimolar concentration (Figure 2B). When it was treated with 1% SDS and 1 mM ME, its fluorescence intensity was mostly recovered to the level of free dye. Comparison of NIR fluorescence images of the free dye, EGF-ATTO655 in PBS, and EGF-ATTO655 in PBS containing 1% SDS + 1 mM ME also confirmed fluorescence quenching and turn-on of the native and denatured EGF-ATTO655. These data indicate that 3-dimentional disintegration of the quenched conjugate can trigger fluorescence turn-on.
Characterization of EGF-ATTO655. (A) Absorption and (B) fluorescence spectra (λex 600 nm) of free ATTO655, EGF-ATTO655 in PBS, and EGF-ATTO655 in PBS containing 1% SDS + 1 mM ME at the concentration of 5 μM dye equiv., respectively. The absorption spectrum of unlabeled EGF is shown for comparison. Inset images: merged images of the bright-field and fluorescence (λex 620/20 nm, λem 670/40 nm) of the sample tubes containing 1) free ATTO655-COOH, 2) EGF-ATTO655 in PBS, and 3) EGF-ATTO655 in PBS containing 1% SDS + 1 mM ME. (C) Stability of quenched state of free dye and EGF-ATTO655 in PBS and PBS containing 10% FBS. Fluorescence intensities of the sample solutions (λex 600 nm, λem 684 nm) were measured for 18 h. (D) Fluorescence turn-on of EGF-ATTO655 upon enzyme treatment. EGF-ATTO655 was reacted with 20 μg/mL proteinase K or 5 mM DTT, and then changes in its fluorescence intensities were measured every 30 min for 18 h.
Since the number of conjugated dyes per an EGF was only 0.78, the fluorescence quenching is likely occurred by PET interaction between ATTO655 and Trp (and/or Tyr), and not by fӧrster resonance energy transfer (FRET) between the conjugated dyes. PET interaction is known to occur very effectively when the fluorophore and the quenchers (here, Trp and Tyr) are located at length scales below 1 nm. Since fluorescence quenching and dequenching of the dye is highly dependent on the proximity of quencher molecules in 1 nm scale, PET-based probes is a sensitive tool for the analysis of conformational changes in small biomolecules such as peptides and nucleotides [23]. Therefore, denaturation of 3-dimensional structure of EGF-ATTO655 with 1% SDS and 1 mM ME caused complete recovery in the fluorescence of the dye. As ATTO655 and Trp (and Tyr) have hydrophobic moieties, they likely tend to form complexes in aqueous solution, thereby resulting in the broadening of UV/Vis absorption spectrum (Figure 2A) and efficient fluorescence quenching of the conjugated dye. Treatment of EGF-ATTO655 with 1% SDS and 1 mM ME distance ATTO655 from the quencher amino acids (Trp and Tyr), and recover UV/Vis absorption spectrum as well as fluorescence emission of the dye. Currently developed PET quenching-based ATTO655 conjugates have advantages compared with the previously reported FRET-based quenched EGF probe [26]: (1) EGF have two lysine residues for conjugation. Therefore, conjugation of one Cy5.5 and one blackhole quencher (BHQ) in an EGF is difficult to control. In contrast, currently developed PET-based probe uses Trp and Tyr in EGF as a quencher, and therefore we don't need to additionally conjugate quenchers to the lysine residue. This not only makes the synthetic step much simpler to perform but also makes conjugation process easy to control. (2) FRET-based EGF probe shows long-distance quenching between Cy5.5 and BHQ, therefore EGF probe must be completely degraded by enzymes for the recovery of quenched fluorescence. However, PET-based EGF probe is highly sensitive to the small changes in distance between the NIR fluorophore and amino acid quenchers, and therefore distance changes between the dye and quencher by 3-dimensional conformation in addition to enzymatic degradation of EGF probe could result in fluorescence turn-on, making the fluorescence recovery more effective.
Stability of quenched state of EGF-ATTO655 in serum condition
Since PET interaction between ATTO655 and Trp/Tyr may be affected in the presence of serum components, we tested the stability of quenched state of EGF-ATTO655 in PBS (pH 7.4) and PBS containing 10% FBS. As a result, no appreciable increase in the fluorescence intensities of the conjugate was observed over the 18 h incubation period in both PBS and serum-contained solutions, indicating that the conjugate is quite stable at physiological pH condition and in the presence of serum proteins (Figure 2C). As mentioned above, PET quenching is highly sensitive in the small distance change between the fluorophore and amino acid quencher. Therefore, no significant changes in fluorescence intensities of EGF-ATTO655 means that distance between ATTO655 dye and Trp (and Tyr) was quite stably maintained at physiological pH condition and in the presence of serum. According to previous reports, use of zwitterionic fluorophores for peptide ligands is advantageous in preventing non-specific binding (with serum proteins) and cell uptake whereas the peptides conjugated with negatively charged near-infrared fluorophores (e.g., IRDye800CW, Cy5.5, and ICG) are prone to non-specific uptake in normal tissues [22, 27]. That seems the reason why PET interaction between ATTO655 and Trp/Tyr wasn't interrupted by serum proteins.
Fluorescence dequenching of EGF-ATTO655 upon enzyme treatment
It is known that upon binding of EGF to its receptor, the EGF-EGFR complexes internalize into the endosomes and these vesicles fuse with lysosomes, where the EGF and EGFR are degraded [28-30]. Therefore, we also tested change in fluorescence intensities upon enzymatic degradation of the EGF (Figure 2D). At first, we checked effect of reducing agents on the quenched status of the conjugate. It is well known that intracellular concentration of glutathione in cancer cells (2- 10 mM) is much higher than that of the extracelluar space (~ 2 μM) [31]. Therefore, we treated the conjugate with reducing agent DTT at 5 mM to mimic intracellular reducing condition, and then fluorescence changes were monitored. As a result, no significant change in fluorescence was observed for 18 h. This data means that reduction of disulfide bonds in EGF did not affect the distance between ATTO655 dye and amino acid quenchers (Trp and Tyr) and therefore didn't induce fluorescence dequenching of EGF-ATTO655. Next, we treated the conjugate with proteinase K for enzymatic degradation of EGF. Proteinase K is known to cleave peptide bonds adjacent to carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups [32]. Therefore, we selected proteinase K as typical endopeptidase with broad specificity for mimicking enzymatic degradation of the conjugate inside the cells (especially in lysosomes). As shown in Figure 2D, fluorescence of quenched EGF-ATTO655 was rapidly increased upon treatment with proteinase K and fully recovered to the level of the free dye at equivalent concentration, supporting that the changes in the distance between the dye and amino acid quenchers by enzymatic degradation also induce complete turn-on of the conjugate's fluorescence. As shown in Figure S3, the fluorescence emission of the free ATTO655 dye was not affected when incubated at physiological and lysosomal pH conditions (i.e., pH 7.4 and 5.0) for over 17 h.
EGFR-targeting specificity of EGF-ATTO655
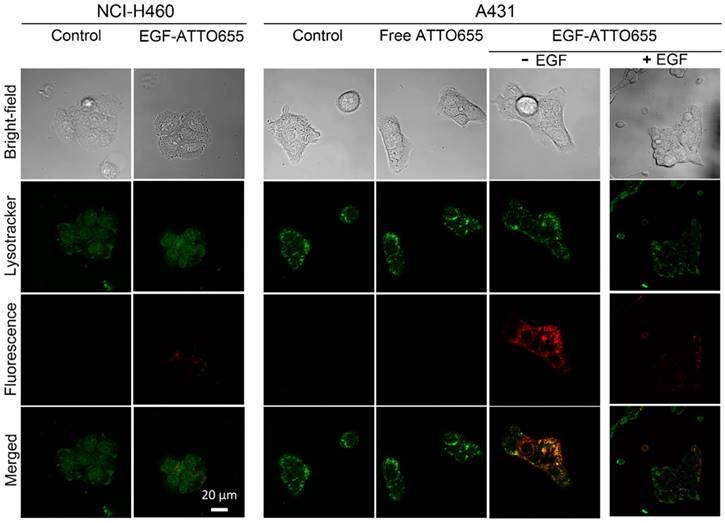
Prior to in vivo study, we evaluated the targeting specificity of EGF-ATTO655 to EGFR-positive cancer cells and subsequent fluorescence turn-on inside the target cells in the in vitro cell imaging study. Both EGFR-positive A431 and EGFR-negative NCI-H460 cancer cells were treated with the conjugate or free dye for 5 h, washed three times, stained with Lysotracker, and then NIR fluorescence images of the both cell lines were obtained by confocal laser scanning microscope (Lysotracker: λex 405 nm, λem 410-585 nm, ATTO655: λex 633 nm, λem 638-759 nm). As shown in Figure 3, strong fluorescence signals of EGF-ATTO655 were observed inside the EGFR-positive A431 cells whereas minor fluorescence was seen in the EGFR-negative NCI-H460 cells. No detectable fluorescence was shown in the free dye-treated A431 cells even after 5 h incubation, indicating that zwitterionic dye itself do not affect non-specific uptake to cancer cells. When A431 cells were pre-treated with the excess amount of unlabeled EGF for competition of receptor binding and then treated with EGF-ATTO655, fluorescence signals were greatly reduced. These data confirm high target-specificity of EGF-ATTO655 to EGFR-positive cancer cells, and subsequent turn-on its fluorescence inside the cells. Bright yellow color in the merged fluorescence image of the Lysotracker and EGF-ATTO655 in A431 cells at 5 h indicates that the conjugate entered into the cells through receptor-mediated endocytosis and preferentially accumulated in the lysosomal sites, where activities of various enzymes as well as concentration of glutathione are high enough to disintegrate the conjugates.
In the meantime, bright fluorescence signals were also detected on the surface of the EGF-ATTO655-treated A431 cells at 1 h post-treatment (Figure S4). This may indicate that the binding of the conjugate to EGFR causes slight changes in the three-dimensional conformation of the dye-conjugated C-terminal region with high structural plasticity (Figure S2), and the changes in the distance between the conjugated dye and amino acid quenchers in the bound conjugate induced the fluorescence recovery of the conjugate to some degree. According to Figure 2B, changes in the 3-dimentional structure of the conjugate with 1% SDS and 1 mM ME induced complete recovery of the quenched fluorescence.
The target specificity of EGF-ATTO655 was also confirmed in the HCC827 (human lung cancer) and HCC70 (human breast cancer) cell lines with moderate EGFR expression levels (Figure S5).
Confocal images of EGF-ATTO655-treated NCI-H460 (EGFR-negative) and A431 (EGFR-positive) cells. The cells were treated with free dye or EGF-ATTO655 for 5 h, and after washing the cells, fluorescence images of the cells were obtained. For competition assay, A431 cells were pre-incubated with excess unlabeled EGF (+ EGF group) and then treated with EGF-ATTO655. Yellow colored regions indicate localization of EGF-ATTO655 in lysosomal sites.
Real-time fluorescence imaging of EGFR-positive cancer cells
Fluorescence turn-on of EGF-ATTO655 inside the target cancer cells was validated in the real-time fluorescence imaging study. EGFR-positive A431cells were treated with free dye and EGF-ATTO655 at the equimolar dye concentration. Since fluorescence of EGF-ATTO655 is expected to be turned on in the targeted cells while the free dye generates bright fluorescence both outside and inside of the cells, NIR fluorescence images of the cancer cells (λex 640/30 nm, λem 690/50 nm) were acquired in real-time without washing steps. As shown in Figure 4 (also see Movie S1 and S2), strong fluorescence signals were observed in the free dye-treated cells for 6 h, and no fluorescence was detected after washing. This means that strong fluorescence was generated by the free dyes in the extracelluar region, and due to the bright background signals, the location of the cancer cells could not be discriminated from the fluorescence images. As shown in Figure 2C, quenched state of the conjugate could be stably maintained for at least 17 h in serum condition at physiological pH 7.4. As a result, minor fluorescence signals were detected in the EGF-ATTO655-treated cells, and fluorescence signals in the extracellular region did not increase for 6 h, confirming the stability of EGF-ATTO655 when located in the outside regions of the target cells. Fluorescence intensities in A431 cells were gradually increased with time, and the strong fluorescence signals from the cells did not disappear after washing step. These data confirm that quenched state of the conjugate could be stably maintained outside the cells and, upon receptor-mediated endocytosis, its fluorescence could be turned on inside the target cells, thereby enabling real-time detection of EGFR-positive cancer cells with high contrast.
According to a previous report [21], fluorescence quenching of Alexa647 by tryptophan and tyrosine is negligible. Therefore, we synthesized EGF-Alexa647 as an always-on type control probe for comparison (see supporting information and Figure S6). After treating A431 cells with EGF-Alexa647, NIR fluorescence images of the cancer cells (λex 640/30 nm, λem 690/50 nm) were acquired in real-time without washing steps. As shown in Figure S7 and Movie S3, strong fluorescence signals were observed for 6 h due to the continuous generation of strong fluorescence from EGF-Alexa647 in the extracellular region. The cells could be discriminated only after the washing step, confirming that strong fluorescence signals generated from the extracellular region before the washing step interrupted the visualization of the EGFR-positive cancer cells in the NIR fluorescence images.
Real-time fluorescence imaging of EGFR-positive A431 cells. The cells were incubated with cell culture medium containing free dye or EGF-ATTO655, and then, without washing steps, fluorescence images of the cells were obtained every 15 min for 6 h. For comparison, the cells were washed at 6 h, and then their fluorescence images were acquired once more.

In vivo and ex vivo NIR fluorescence imaging
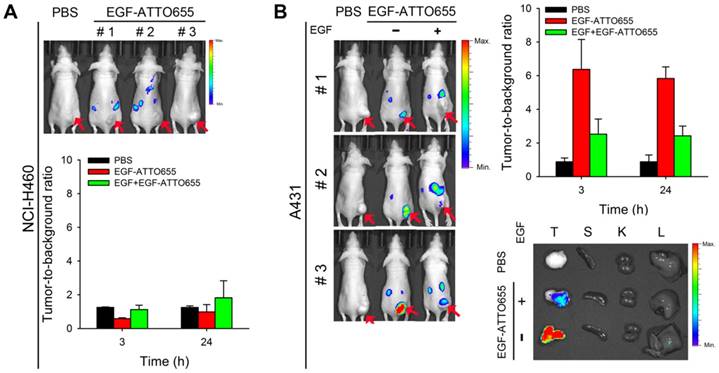
For the evaluation of EGF-ATTO655 for target-specific imaging of EGFR-overexpressing tumors, systemic injection of EGF-ATTO655 was applied to the mice bearing A431 (EGFR-positive) and NCI-H460 (EGFR-negative) tumors, and then NIR fluorescence images of the mice were obtained using an IVIS Lumina imaging system at 3 and 24 h post-injection (λex = 620/20 nm, λem = 670/40 nm), and their TBRs at different time points were analyzed. According to Figure 5A, no appreciable fluorescence signals in tumor sites compared with the surrounding normal tissues were detected in the NCI-H460 tumor-bearing mice, resulting in no enhancement in TBR compared with PBS control group (Figure 5). In contrast, the tumor sites of A431 tumor-bearing mice could be clearly identified from the fluorescence images with high TBR (that is, 6.37 ± 1.78 at 3 h and 5.83 ± 0.69 at 24 post-injection). Tumor-specificity of the conjugate was also verified by co-injecting the excess amount of unlabeled EGF with EGF-ATTO655 into the A431 tumor-bearing mice. As shown in Figure 5B, significantly reduced fluorescence intensities in tumors and lowered TBR value confirm tumor-specificity of the conjugate. Ex vivo images of the tumors and organs indicates accumulation of the conjugate and subsequent turn-on of its NIR fluorescence were mainly occurred in the EGFR-positive tumors.
As another control experiment, the mice bearing EGFR-positive A431 tumors (n=4) received an intravenous injection of EGF-Alexa647; then, NIR fluorescence images of the mice were obtained using an IVIS Lumina imaging system at 3 and 24 h post-injection (λex = 620/20 nm, λem = 670/40 nm), and their TBRs at different time points were analyzed. As shown in Figure S9A and B, strong fluorescence signals were generated from the circulating EGF-Alexa647, thereby keeping the background signals high for 24 h. As expected, the TBR values analyzed at 3 and 24 h post-injection were only 2.44 ± 0.13 and 2.25 ± 0.57, respectively. In particular, from the ex vivo imaging of the collected organs, it was observed that the mean fluorescence intensity from the kidney was 4.85 times higher than that from the tumor (Figure S9C). High fluorescence signals from the kidneys may be due to both high EGFR expression in the liver and kidneys, and renal elimination of the small-sized EGF-fluorophore conjugates [34].
In vivo NIR fluorescence imaging of EGFR-positive tumors. (A) NIR fluorescence images of the mice bearing EGFR-negative NCI-H460 tumors at 3 h post-injection of EGF-ATTO655 (upper panel), and analysis of tumor-to-background ratio (TBR) at different time points (lower. (B) NIR fluorescence images of the mice bearing EGFR-positive A431 tumors at 3 h post-injection of EGF-ATTO655 (left panel), and analysis of tumor-to-background ratio (TBR) at different time points (right panel). Results from ex vivo imaging of the tumor and organs (T: tumor, S: spleen, K: kidney, L: liver) are also shown. The tumor-bearing mice with (+ EGF) and without (- EGF) co-treatment of excess unlabeled EGF are compared. Arrows indicate tumor sites.
Confocal fluorescence microscopy images of tumor sections. Nuclei of the cells in the tumor sections were counterstained with DAPI. The merged fluorescence images of DAPI (λex 405 nm, λem 410-482 nm) and EGF-ATTO655 (λex 633 nm, λem 638-758 nm).
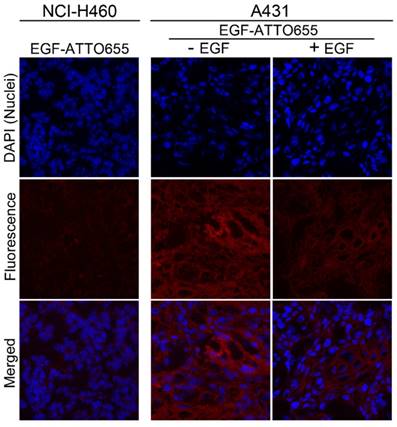
Finally, intratumoral distribution of EGF-ATTO655 was evaluated with tumor sections. As shown in Figure 6, no measurable fluorescence was observed in the NCI-H460 tumor section whereas high fluorescence signals were detected in A431 tumor section. And also, fluorescence signals were remarkably reduced when the excess unlabeled EGF was co-treated with EGF-ATTO655. This result confirms that EGF-ATTO655 specifically accumulated in EGFR-overexpressed tumors and its fluorescence was turned on in the tumor cells.
Of note is that fluorescence signals of EGF-ATTO655 in the tumor sections were broadly distributed which means that this relatively small-sized conjugate (< 7 kDa) could easily penetrate into the deep sites of the solid tumor mass whereas large sized antibody-NIR dye conjugates (~ 150 kDa) have poor tissue permeability [9-11, 26, 33].
Notably, we showed that average TBR in the A431 tumor-bearing mice treated with EGF-ATTO655 was 6.37 at 3 h post-injection. This is quite an interesting result because in the case of EGFR-target specific cetuximab-IRDye800CW, calculated TBR was approximately 3 in the animal study after 2 days of intravenous injection [12]. In addition, in the clinical trials of cetuximab-IRDye800CW, fluorescence image-guided imaging and surgery is being carried out after 2 days of systemic injection of the conjugate, and observed TBR in patients were ~ 4 [14]. The high TBR value obtained in EGF-ATTO655 conjugate was due to the combinations of specific fluorescence turn-on of NIR fluorescence in target cells, fast tumor localization, and rapid excretion of the small sized EGF-ATTO655 through the kidneys [34, 35]. These kinds of results could not be obtained by using the always-on type EGF-fluorophore conjugates (e.g., EGF-Alexa647). Therefore, PET-based EGF-ATTO655 probe is shown to have great potential for fast and target-specific NIR fluorescence imaging of EGFR-positive tumors in vivo.
Conclusions
We developed a PET-based zwitterionic fluorophore-conjugated EGF probe for fast, real-time, and target-cell-specific NIR fluorescence imaging of EGFR-positive cancers in vitro and in vivo. The NIR fluorescence of EGF-ATTO655 was remarkably quenched via PET interaction between the conjugated ATTO655 dye and amino acid quenchers (Trp and Tyr) in EGF, and its quenched state was stably maintained at physiological pH and in the presence of serum for at least 17 h. Fluorescence of the conjugate could be turned on after changes in 3-dimensional structure and/or enzymatic degradation in the target cells, thereby enabling fast and real-time fluorescence identification of EGFR-positive cancer cells. Since this small-sized EGF-ATTO655 has advantageous characteristics, including OFF/ON optical property, rapid clearance from the body thorough urinary excretion, and good tissue permeability, EGFR-positive tumors in animal studies could be clearly visualized with high TBR within 3 h post-injection. Therefore, this PET interaction-based OFF/ON type of EGF probe may have great potential for fast, real-time, and target-cell-specific imaging of EGFR-positive cancers.
Supplementary Material
Supplementary methods, figures and tables.
Supplementary movie 1 - Free dye.
Supplementary movie 2 - EGF-ATTO655.
Supplementary movie 3 - EGF-Alexa647.
Acknowledgements
This work was supported by the National Cancer Center grant (1610150, 1910070 and 1811020), and the Ministry of Oceans and Fisheries (project title: “Development of marine material based near infrared fluorophore complex and diagnostic imaging instruments), Korea. We thank the Proteomics Core Facility at the National Cancer Center in Korea, which provided mass spectrometry services.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ferguson KM, Darling PJ, Mohan MJ, Macatee TL, Lemmon MA. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. EMBO J. 2000;19:4632-43
2. Kari C, Chan TO, Rocha de Quadros M, Rodeck U. Targeting the epidermal growth factor receptor in cancer: apoptosis takes center stage. Cancer Res. 2003;63:1-5
3. Yarden Y, Sliwkowski MX. Untangling the erbB signaling network. Nat Rev Mol Cell Biol. 2001;2:127-37
4. Baselga J. The EGFR as a target for anticancer therapy-focus on cetuximab. Eur J Cancer. 2001;37:16-22
5. Ciardiello F, Tortor G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160-74
6. Teicher BA. Antibody-drug conjugate targets. Curr Cancer Drug Targets. 2009;9:982-1004
7. Baselga J. Targeting tyrosine kinases in cancer: the second wave. Science. 2006;312:1175-8
8. Speake G, Holloway B, Costello G. Recent developments related to the EGFR as a target for cancer chemotherapy. Curr Opin Pharmacol. 2005;5:343-9
9. Image guided surgery for margin assessment of head and neck cancer using cetuximab-IRDye800CW conjugate, 11 January 2018. https://clinicaltrials.gov/ct2/show/NCT03134846?cond=IMAGE+GUIDED+SURGERY+FOR+MARGIN+ASSESSMENT&rank=1
10. Cetuximab-IRDye 800CW and intraoperative imaging in finding pancreatic cancer in patients undergoing surgery, 18 June 2018. https://clinicaltrials.gov/ct2/show/NCT02736578?cond=NCT02736578&rank=1
11. Cetuximab-IRDye800CW in detecting tumors in patients with malignant glioma undergoing surgery, 14 May 2018. https://clinicaltrials.gov/ct2/show/NCT02855086?cond=NCT02855086&rank=1
12. Day KE, Sweeny L, Kulbersh B, Zinn KR, Rosenthal EL. Preclinical comparison of near-infrared-labeled cetuximab and panitumumab for optical imaging of head and neck squamous cell carcinoma. Mol Imaging Biol. 2013;15:722-9
13. Rosenthal EL, Moore LS, Tipirneni K. et al. Sensitivity and specificity of cetuximab-IRDye800CW to identify regional metastatic disease in head and neck cancer. Clin Cancer Res. 2017;23:4744-52
14. Miller E, Tummers WS, Teraphongphom N. et al. First-in-human intraoperative near-infrared fluorescence imaging of glioblastoma using cetuximab-IRDye800. J Neurooncol. 2018;139:135-43
15. Choi HS, Gibbs SL, Lee JH. et al. Targeted zwitterionic near-infrared fluorophores for improved optical imaging. Nat Biotechnol. 2013;31:148-53
16. Diagaradjane P, Orenstein-Cardona JM, Colón-Casasnovas NE. et al. Imaging epidermal growth factor receptor expression in vivo: pharmacokinetic and biodistribution characterization of a bioconjugated quantum dot nanoprobe. Clin Cancer Res. 2008;14:731-41
17. Thomas TP, Shukla R, Kotlyar A. et al. Dendrimer-epidermal growth factor conjugate displays superagonist activity. Biomacromolecules. 2008;9:603-9
18. Creixell M, Bohórquez AC, Torres-Lugo M, Rinaldi C. EGFR-targeted magnetic nanoparticle heaters kill cancer cells without a perceptible temperature rise. ACS Nano. 2011;5:7124-9
19. Jones JT, Akita RW, Sliwkowski MX. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 1999;447:227-31
20. Carpenter G, Cohen S. Epidermal growth factor. J Biol Chem. 1990;265:7709-12
21. Marmé N, Knemeyer JP, Sauer M, Wolfrum J. Inter- and intramolecular fluorescence quenching of organic dyes by tryptophan. Bioconjugate Chem. 2003;14:1133-9
22. Choi HS, Gibbs SL, Lee JH. et al. Targeted zwitterionic near-infrared fluorophores for improved optical imaging. Nat Biotechnol. 2013;31:148-53
23. Doose S, Neuweiler H, Sauer M, Close A. Look at fluorescence quenching of organic dyes by tryptophan. ChemPhysChem. 2005;6:2277-85
24. Kim JC, Ali MA, Nandi A. et al. Correlation of HER1/EGFR expression and degree of radiosensitizing effect of the HER1/EGFR-tyrosine kinase inhibitor erlotinib. Indian J Biochem Biophys. 2005;42:358-65
25. Dong A, Wodzlak D, Lowe AW. Epidermal growth factor receptor (EGFR) signaling requires a specific endoplasmic reticulum thioredoxin for the post-translational control of receptor presentation to the cell surface. J Biol Chem. 2015;290:8016-27
26. Ryu JH, Shin M, Kim SA. et al. In vivo fluorescence imaging for cancer diagnosis using receptor-targeted epidermal growth factor-based nanoprobe. Biomaterials. 2013;34:9149-59
27. Choi HS, Liu W, Misra P. et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165-70
28. Schlessinger J, Schechter Y, Willingham MC, Pnstan I. Direct visualization of binding, aggregation and internalization of insulin and epidermal growth factor in living fibroblastic cells. Proc Natl Acad Sci U S A. 1978;75:2659-63
29. Gorden P, Carpentier JL, Cohen S, Orci L. Epidermal growth factor: morphological demonstration of binding, internalization, and lysosomal association in human fibroblasts. Proc Natl Acad Sci U S A. 1978;75:5025-9
30. Stoscheck CM, Carpenter G. Characterization of the metabolic turnover of epidermal growth factor receptor protein in A431 cells. J Cell Physiol. 1984;120:296-302
31. Estrela JM, Ortega A, Obrador E. Glutathione in cancer biology and therapy. Crit Rev Clin Lab Sci. 2006;43:143-81
32. Morihara K, Tsuzuki H. Specificity of proteinase K from Tritirachium album Limber for synthetic peptides. Agric Biol Chem. 1975;39:1489-92
33. Gong H, Kovar J, Little G, Chen H, Olive DM. In vivo imaging of xenograft tumors using an epidermal growth factor receptor-specific affibody molecule labeled with a near-infrared fluorophore. Neoplasia. 2010;12:139-49
34. Reilly RM, Kiarash R, Sandhu J. et al. A comparison of EGF and MAb 528 labeled with 111In for imaging human breast cancer. J Nucl Med. 2000;41:903-11
35. Jia L, Zhang L, Shao C. et al. An attempt to understand kidney's protein handling function by comparing plasma and urine proteomes. PLoS One. 2009;4:e5146
Author contact
![]() Corresponding author: Yongdoo Choi, Ph.D. Tel: +82-31-920-2512; E-mail: ydchoire.kr
Corresponding author: Yongdoo Choi, Ph.D. Tel: +82-31-920-2512; E-mail: ydchoire.kr
Received 2018-9-5
Accepted 2018-12-26
Published 2019-1-30
 Global reach, higher impact
Global reach, higher impact