Metalloprotein
Metalloprotein is a generic term for a protein that contains a metal ion cofactor.[1][2] A large proportion of all proteins are part of this category. For instance, at least 1000 human proteins (out of ~20,000) contain zinc-binding protein domains[3] although there may be up to 3000 human zinc metalloproteins.[4]
Abundance
[edit]It is estimated that approximately half of all proteins contain a metal.[5] In another estimate, about one quarter to one third of all proteins are proposed to require metals to carry out their functions.[6] Thus, metalloproteins have many different functions in cells, such as storage and transport of proteins, enzymes and signal transduction proteins, or infectious diseases.[7] The abundance of metal binding proteins may be inherent to the amino acids that proteins use, as even artificial proteins without evolutionary history will readily bind metals.[8]
Most metals in the human body are bound to proteins. For instance, the relatively high concentration of iron in the human body is mostly due to the iron in hemoglobin.
| Liver | Kidney | Lung | Heart | Brain | Muscle | |
|---|---|---|---|---|---|---|
| Mn (manganese) | 138 | 79 | 29 | 27 | 22 | <4-40 |
| Fe (iron) | 16,769 | 7,168 | 24,967 | 5,530 | 4,100 | 3,500 |
| Co (cobalt) | <2-13 | <2 | <2-8 | --- | <2 | 150 (?) |
| Ni (nickel) | <5 | <5-12 | <5 | <5 | <5 | <15 |
| Cu (copper) | 882 | 379 | 220 | 350 | 401 | 85-305 |
| Zn (zinc) | 5,543 | 5,018 | 1,470 | 2,772 | 915 | 4,688 |
Coordination chemistry principles
[edit]In metalloproteins, metal ions are usually coordinated by nitrogen, oxygen or sulfur centers belonging to amino acid residues of the protein. These donor groups are often provided by side-chains on the amino acid residues. Especially important are the imidazole substituent in histidine residues, thiolate substituents in cysteine residues, and carboxylate groups provided by aspartate. Given the diversity of the metalloproteome, virtually all amino acid residues have been shown to bind metal centers. The peptide backbone also provides donor groups; these include deprotonated amides and the amide carbonyl oxygen centers. Lead(II) binding in natural and artificial proteins has been reviewed.[10]
In addition to donor groups that are provided by amino acid residues, many organic cofactors function as ligands. Perhaps most famous are the tetradentate N4 macrocyclic ligands incorporated into the heme protein. Inorganic ligands such as sulfide and oxide are also common.
Storage and transport metalloproteins
[edit]These are the second stage product of protein hydrolysis obtained by treatment with slightly stronger acids and alkalies.
Oxygen carriers
[edit]Hemoglobin, which is the principal oxygen-carrier in humans, has four subunits in which the iron(II) ion is coordinated by the planar macrocyclic ligand protoporphyrin IX (PIX) and the imidazole nitrogen atom of a histidine residue. The sixth coordination site contains a water molecule or a dioxygen molecule. By contrast the protein myoglobin, found in muscle cells, has only one such unit. The active site is located in a hydrophobic pocket. This is important as without it the iron(II) would be irreversibly oxidized to iron(III). The equilibrium constant for the formation of HbO2 is such that oxygen is taken up or released depending on the partial pressure of oxygen in the lungs or in muscle. In hemoglobin the four subunits show a cooperativity effect that allows for easy oxygen transfer from hemoglobin to myoglobin.[11]
In both hemoglobin and myoglobin it is sometimes incorrectly stated that the oxygenated species contains iron(III). It is now known that the diamagnetic nature of these species is because the iron(II) atom is in the low-spin state. In oxyhemoglobin the iron atom is located in the plane of the porphyrin ring, but in the paramagnetic deoxyhemoglobin the iron atom lies above the plane of the ring.[11] This change in spin state is a cooperative effect due to the higher crystal field splitting and smaller ionic radius of Fe2+ in the oxyhemoglobin moiety.
Hemerythrin is another iron-containing oxygen carrier. The oxygen binding site is a binuclear iron center. The iron atoms are coordinated to the protein through the carboxylate side chains of a glutamate and aspartate and five histidine residues. The uptake of O2 by hemerythrin is accompanied by two-electron oxidation of the reduced binuclear center to produce bound peroxide (OOH−). The mechanism of oxygen uptake and release have been worked out in detail.[12][13]
Hemocyanins carry oxygen in the blood of most mollusks, and some arthropods such as the horseshoe crab. They are second only to hemoglobin in biological popularity of use in oxygen transport. On oxygenation the two copper(I) atoms at the active site are oxidized to copper(II) and the dioxygen molecules are reduced to peroxide, O2−
2.[14][15]
Chlorocruorin (as the larger carrier erythrocruorin) is an oxygen-binding hemeprotein present in the blood plasma of many annelids, particularly certain marine polychaetes.
Cytochromes
[edit]Oxidation and reduction reactions are not common in organic chemistry as few organic molecules can act as oxidizing or reducing agents. Iron(II), on the other hand, can easily be oxidized to iron(III). This functionality is used in cytochromes, which function as electron-transfer vectors. The presence of the metal ion allows metalloenzymes to perform functions such as redox reactions that cannot easily be performed by the limited set of functional groups found in amino acids.[16] The iron atom in most cytochromes is contained in a heme group. The differences between those cytochromes lies in the different side-chains. For instance cytochrome a has a heme a prosthetic group and cytochrome b has a heme b prosthetic group. These differences result in different Fe2+/Fe3+ redox potentials such that various cytochromes are involved in the mitochondrial electron transport chain.[17]
Cytochrome P450 enzymes perform the function of inserting an oxygen atom into a C−H bond, an oxidation reaction.[18][19]
Rubredoxin
[edit]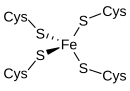
Rubredoxin is an electron-carrier found in sulfur-metabolizing bacteria and archaea. The active site contains an iron ion coordinated by the sulfur atoms of four cysteine residues forming an almost regular tetrahedron. Rubredoxins perform one-electron transfer processes. The oxidation state of the iron atom changes between the +2 and +3 states. In both oxidation states the metal is high spin, which helps to minimize structural changes.
Plastocyanin
[edit]
Plastocyanin is one of the family of blue copper proteins that are involved in electron transfer reactions. The copper-binding site is described as distorted trigonal pyramidal.[20] The trigonal plane of the pyramidal base is composed of two nitrogen atoms (N1 and N2) from separate histidines and a sulfur (S1) from a cysteine. Sulfur (S2) from an axial methionine forms the apex. The distortion occurs in the bond lengths between the copper and sulfur ligands. The Cu−S1 contact is shorter (207 pm) than Cu−S2 (282 pm). The elongated Cu−S2 bonding destabilizes the Cu(II) form and increases the redox potential of the protein. The blue color (597 nm peak absorption) is due to the Cu−S1 bond where S(pπ) to Cu(dx2−y2) charge transfer occurs.[21]
In the reduced form of plastocyanin, His-87 will become protonated with a pKa of 4.4. Protonation prevents it acting as a ligand and the copper site geometry becomes trigonal planar.
Metal-ion storage and transfer
[edit]Iron
[edit]Iron is stored as iron(III) in ferritin. The exact nature of the binding site has not yet been determined. The iron appears to be present as a hydrolysis product such as FeO(OH). Iron is transported by transferrin whose binding site consists of two tyrosines, one aspartic acid and one histidine.[22] The human body has no controlled mechanism for excretion of iron.[23] This can lead to iron overload problems in patients treated with blood transfusions, as, for instance, with β-thalassemia. Iron is actually excreted in urine[24] and is also concentrated in bile[25] which is excreted in feces.[26]
Copper
[edit]Ceruloplasmin is the major copper-carrying protein in the blood. Ceruloplasmin exhibits oxidase activity, which is associated with possible oxidation of Fe(II) into Fe(III), therefore assisting in its transport in the blood plasma in association with transferrin, which can carry iron only in the Fe(III) state.
Calcium
[edit]Osteopontin is involved in mineralization in the extracellular matrices of bones and teeth.
Metalloenzymes
[edit]Metalloenzymes all have one feature in common, namely that the metal ion is bound to the protein with one labile coordination site. As with all enzymes, the shape of the active site is crucial. The metal ion is usually located in a pocket whose shape fits the substrate. The metal ion catalyzes reactions that are difficult to achieve in organic chemistry.
Carbonic anhydrase
[edit]
In aqueous solution, carbon dioxide forms carbonic acid
- CO2 + H2O ⇌ H2CO3
This reaction is very slow in the absence of a catalyst, but quite fast in the presence of the hydroxide ion
- CO2 + OH− ⇌ HCO−
3
A reaction similar to this is almost instantaneous with carbonic anhydrase. The structure of the active site in carbonic anhydrases is well known from a number of crystal structures. It consists of a zinc ion coordinated by three imidazole nitrogen atoms from three histidine units. The fourth coordination site is occupied by a water molecule. The coordination sphere of the zinc ion is approximately tetrahedral. The positively-charged zinc ion polarizes the coordinated water molecule, and nucleophilic attack by the negatively-charged hydroxide portion on carbon dioxide proceeds rapidly. The catalytic cycle produces the bicarbonate ion and the hydrogen ion[2] as the equilibrium:
- H2CO3 ⇌ HCO−
3 + H+
favouring dissociation of carbonic acid at biological pH values.[27]
Vitamin B12-dependent enzymes
[edit]The cobalt-containing Vitamin B12 (also known as cobalamin) catalyzes the transfer of methyl (−CH3) groups between two molecules, which involves the breaking of C−C bonds, a process that is energetically expensive in organic reactions. The metal ion lowers the activation energy for the process by forming a transient Co−CH3 bond.[28] The structure of the coenzyme was famously determined by Dorothy Hodgkin and co-workers, for which she received a Nobel Prize in Chemistry.[29] It consists of a cobalt(II) ion coordinated to four nitrogen atoms of a corrin ring and a fifth nitrogen atom from an imidazole group. In the resting state there is a Co−C sigma bond with the 5′ carbon atom of adenosine.[30] This is a naturally occurring organometallic compound, which explains its function in trans-methylation reactions, such as the reaction carried out by methionine synthase.
Nitrogenase (nitrogen fixation)
[edit]The fixation of atmospheric nitrogen is an energy-intensive process, as it involves breaking the very stable triple bond between the nitrogen atoms. The nitrogenases catalyze the process. One such enzyme occurs in Rhizobium bacteria. There are three components to its action: a molybdenum atom at the active site, iron–sulfur clusters that are involved in transporting the electrons needed to reduce the nitrogen, and an abundant energy source in the form of magnesium ATP. This last is provided by a mutualistic symbiosis between the bacteria and a host plant, often a legume. The reaction may be written symbolically as
where Pi stands for inorganic phosphate. The precise structure of the active site has been difficult to determine. It appears to contain a MoFe7S8 cluster that is able to bind the dinitrogen molecule and, presumably, enable the reduction process to begin.[31] The electrons are transported by the associated "P" cluster, which contains two cubical Fe4S4 clusters joined by sulfur bridges.[32]
Superoxide dismutase
[edit]
The superoxide ion, O−
2 is generated in biological systems by reduction of molecular oxygen. It has an unpaired electron, so it behaves as a free radical. It is a powerful oxidizing agent. These properties render the superoxide ion very toxic and are deployed to advantage by phagocytes to kill invading microorganisms. Otherwise, the superoxide ion must be destroyed before it does unwanted damage in a cell. The superoxide dismutase enzymes perform this function very efficiently.[33]
The formal oxidation state of the oxygen atoms is −1⁄2. In solutions at neutral pH, the superoxide ion disproportionates to molecular oxygen and hydrogen peroxide.
- 2 O−
2 + 2 H+ → O2 + H2O2
In biology this type of reaction is called a dismutation reaction. It involves both oxidation and reduction of superoxide ions. The superoxide dismutase (SOD) group of enzymes increase the rate of reaction to near the diffusion-limited rate.[34] The key to the action of these enzymes is a metal ion with variable oxidation state that can act either as an oxidizing agent or as a reducing agent.
- Oxidation: M(n+1)+ + O−
2 → Mn+ + O2 - Reduction: Mn+ + O−
2 + 2 H+ → M(n+1)+ + H2O2.
In human SOD, the active metal is copper, as Cu(II) or Cu(I), coordinated tetrahedrally by four histidine residues. This enzyme also contains zinc ions for stabilization and is activated by copper chaperone for superoxide dismutase (CCS). Other isozymes may contain iron, manganese or nickel. The activity of Ni-SOD involves nickel(III), an unusual oxidation state for this element. The active site nickel geometry cycles from square planar Ni(II), with thiolate (Cys2 and Cys6) and backbone nitrogen (His1 and Cys2) ligands, to square pyramidal Ni(III) with an added axial His1 side chain ligand.[35]
Chlorophyll-containing proteins
[edit]
Chlorophyll plays a crucial role in photosynthesis. It contains a magnesium enclosed in a chlorin ring. However, the magnesium ion is not directly involved in the photosynthetic function and can be replaced by other divalent ions with little loss of activity. Rather, the photon is absorbed by the chlorin ring, whose electronic structure is well-adapted for this purpose.
Initially, the absorption of a photon causes an electron to be excited into a singlet state of the Q band. The excited state undergoes an intersystem crossing from the singlet state to a triplet state in which there are two electrons with parallel spin. This species is, in effect, a free radical, and is very reactive and allows an electron to be transferred to acceptors that are adjacent to the chlorophyll in the chloroplast. In the process chlorophyll is oxidized. Later in the photosynthetic cycle, chlorophyll is reduced back again. This reduction ultimately draws electrons from water, yielding molecular oxygen as a final oxidation product.
Hydrogenase
[edit]Hydrogenases are subclassified into three different types based on the active site metal content: iron–iron hydrogenase, nickel–iron hydrogenase, and iron hydrogenase.[36] All hydrogenases catalyze reversible H2 uptake, but while the [FeFe] and [NiFe] hydrogenases are true redox catalysts, driving H2 oxidation and H+ reduction
- H2 ⇌ 2 H+ + 2 e−
the [Fe] hydrogenases catalyze the reversible heterolytic cleavage of H2.
- H2 ⇌ H+ + H−

Ribozyme and deoxyribozyme
[edit]Since discovery of ribozymes by Thomas Cech and Sidney Altman in the early 1980s, ribozymes have been shown to be a distinct class of metalloenzymes.[37] Many ribozymes require metal ions in their active sites for chemical catalysis; hence they are called metalloenzymes. Additionally, metal ions are essential for structural stabilization of ribozymes. Group I intron is the most studied ribozyme which has three metals participating in catalysis.[38] Other known ribozymes include group II intron, RNase P, and several small viral ribozymes (such as hammerhead, hairpin, HDV, and VS) and the large subunit of ribosomes. Several classes of ribozymes have been described.[39]
Deoxyribozymes, also called DNAzymes or catalytic DNA, are artificial DNA-based catalysts that were first produced in 1994.[40] Almost all DNAzymes require metal ions. Although ribozymes mostly catalyze cleavage of RNA substrates, a variety of reactions can be catalyzed by DNAzymes including RNA/DNA cleavage, RNA/DNA ligation, amino acid phosphorylation and dephosphorylation, and carbon–carbon bond formation.[41] Yet, DNAzymes that catalyze RNA cleavage reaction are the most extensively explored ones. 10-23 DNAzyme, discovered in 1997, is one of the most studied catalytic DNAs with clinical applications as a therapeutic agent.[42] Several metal-specific DNAzymes have been reported including the GR-5 DNAzyme (lead-specific),[43] the CA1-3 DNAzymes (copper-specific), the 39E DNAzyme (uranyl-specific)[44] and the NaA43 DNAzyme (sodium-specific).[45]
Signal-transduction metalloproteins
[edit]Calmodulin
[edit]
Calmodulin is an example of a signal-transduction protein. It is a small protein that contains four EF-hand motifs, each of which is able to bind a Ca2+ ion.
In an EF-hand loop protein domain, the calcium ion is coordinated in a pentagonal bipyramidal configuration. Six glutamic acid and aspartic acid residues involved in the binding are in positions 1, 3, 5, 7 and 9 of the polypeptide chain. At position 12, there is a glutamate or aspartate ligand that behaves as a bidentate ligand, providing two oxygen atoms. The ninth residue in the loop is necessarily glycine due to the conformational requirements of the backbone. The coordination sphere of the calcium ion contains only carboxylate oxygen atoms and no nitrogen atoms. This is consistent with the hard nature of the calcium ion.
The protein has two approximately symmetrical domains, separated by a flexible "hinge" region. Binding of calcium causes a conformational change to occur in the protein. Calmodulin participates in an intracellular signaling system by acting as a diffusible second messenger to the initial stimuli.[46][47]
Troponin
[edit]In both cardiac and skeletal muscles, muscular force production is controlled primarily by changes in the intracellular calcium concentration. In general, when calcium rises, the muscles contract and, when calcium falls, the muscles relax. Troponin, along with actin and tropomyosin, is the protein complex to which calcium binds to trigger the production of muscular force.
Transcription factors
[edit]
Many transcription factors contain a structure known as a zinc finger, a structural module in which a region of protein folds around a zinc ion. The zinc does not directly contact the DNA that these proteins bind to. Instead, the cofactor is essential for the stability of the tightly folded protein chain.[48] In these proteins, the zinc ion is usually coordinated by pairs of cysteine and histidine side-chains.
Other metalloenzymes
[edit]There are two types of carbon monoxide dehydrogenase: one contains iron and molybdenum, the other contains iron and nickel. Parallels and differences in catalytic strategies have been reviewed.[49]
Pb2+ (lead) can replace Ca2+ (calcium) as, for example, with calmodulin or Zn2+ (zinc) as with metallocarboxypeptidases.[50]
Some other metalloenzymes are given in the following table, according to the metal involved.
See also
[edit]References
[edit]- ^ Banci L (2013). "Metallomics and the Cell: Some Definitions and General Comments". In Sigel A, Sigel H, Sigel RK (eds.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. pp. 1–13. doi:10.1007/978-94-007-5561-1_1. ISBN 978-94-007-5561-1. PMID 23595668.
- ^ a b Shriver DF, Atkins PW (1999). "Charper 19, Bioinorganic chemistry". Inorganic chemistry (3rd ed.). Oxford University Press. ISBN 978-0-19-850330-9.
- ^ Human reference proteome in Uniprot, accessed 12 Jan 2018
- ^ Andreini C, Banci L, Bertini I, Rosato A (November 2006). "Zinc through the three domains of life". Journal of Proteome Research. 5 (11): 3173–8. doi:10.1021/pr0603699. PMID 17081069.
- ^ Thomson AJ, Gray HB (1998). "Bioinorganic chemistry" (PDF). Current Opinion in Chemical Biology. 2 (2): 155–158. doi:10.1016/S1367-5931(98)80056-2. PMID 9667942.
- ^ Waldron KJ, Robinson NJ (January 2009). "How do bacterial cells ensure that metalloproteins get the correct metal?". Nature Reviews. Microbiology. 7 (1): 25–35. doi:10.1038/nrmicro2057. PMID 19079350. S2CID 7253420.
- ^ Carver PL (2013). "Metal Ions and Infectious Diseases. An Overview from the Clinic". In Sigel A, Sigel H, Sigel RK (eds.). Interrelations between Essential Metal Ions and Human Diseases. Metal Ions in Life Sciences. Vol. 13. Springer. pp. 1–28. doi:10.1007/978-94-007-7500-8_1. ISBN 978-94-007-7499-5. PMID 24470087.
- ^ Wang, MS; Hoegler, KH; Hecht, M (2019). "Unevolved De Novo Proteins Have Innate Tendencies to Bind Transition Metals". Life. 9 (8): 8. Bibcode:2019Life....9....8W. doi:10.3390/life9010008. PMC 6463171. PMID 30634485.
- ^ Maret W (February 2010). "Metalloproteomics, metalloproteomes, and the annotation of metalloproteins". Metallomics. 2 (2): 117–25. doi:10.1039/b915804a. PMID 21069142.
- ^ Cangelosi V, Ruckthong L, Pecoraro VL (2017). "Chapter 10. Lead(II) Binding in Natural and Artificial Proteins". In Astrid S, Helmut S, Sigel RK (eds.). Lead: Its Effects on Environment and Health. Metal Ions in Life Sciences. Vol. 17. de Gruyter. pp. 271–318. doi:10.1515/9783110434330-010. ISBN 9783110434330. PMC 5771651. PMID 28731303.
- ^ a b Greenwood, Norman N.; Earnshaw, Alan (1997). Chemistry of the Elements (2nd ed.). Butterworth-Heinemann. ISBN 978-0-08-037941-8. Fig.25.7, p 1100 illustrates the structure of deoxyhemoglobin
- ^ Stenkamp, R. E. (1994). "Dioxygen and hemerythrin". Chem. Rev. 94 (3): 715–726. doi:10.1021/cr00027a008.
- ^ Wirstam M, Lippard SJ, Friesner RA (April 2003). "Reversible dioxygen binding to hemerythrin". Journal of the American Chemical Society. 125 (13): 3980–7. doi:10.1021/ja017692r. PMID 12656634.
- ^ Karlin K, Cruse RW, Gultneh Y, Farooq A, Hayes JC, Zubieta J (1987). "Dioxygen–copper reactivity. Reversible binding of O2 and CO to a phenoxo-bridged dicopper(I) complex". J. Am. Chem. Soc. 109 (9): 2668–2679. doi:10.1021/ja00243a019.
- ^ Kitajima N, Fujisawa K, Fujimoto C, Morooka Y, Hashimoto S, Kitagawa T, Toriumi K, Tatsumi K, Nakamura A (1992). "A new model for dioxygen binding in hemocyanin. Synthesis, characterization, and molecular structure of the μ-η2:η2-peroxo dinuclear copper(II) complexes, [Cu(Hb(3,5-R2pz)3)]2(O2) (R = isopropyl and Ph)". J. Am. Chem. Soc. 114 (4): 1277–1291. doi:10.1021/ja00030a025.
- ^ Messerschmidt A, Huber R, Wieghardt K, Poulos T (2001). Handbook of Metalloproteins. Wiley. ISBN 978-0-471-62743-2.
- ^ Moore GR, Pettigrew GW (1990). Cytochrome c: Structural and Physicochemical Aspects. Berlin: Springer.
- ^ Sigel A, Sigel H, Sigel RK, eds. (2007). The Ubiquitous Roles of Cytochrome 450 Proteins. Metal Ions in Life Sciences. Vol. 3. Wiley. ISBN 978-0-470-01672-5.
- ^ Ortiz de Montellano P (2005). Cytochrome P450 Structure, Mechanism, and Biochemistry (3rd ed.). Springer. ISBN 978-0-306-48324-0.
- ^ Colman PM, Freeman HC, Guss JM, Murata M, Norris VA, Ramshaw JA, Venkatappa MP (1978). "X-Ray Crystal-Structure Analysis of Plastocyanin at 2.7 Å Resolution". Nature. 272 (5651): 319–324. Bibcode:1978Natur.272..319C. doi:10.1038/272319a0. S2CID 4226644.
- ^ Solomon EI, Gewirth AA, Cohen SL (1986). Spectroscopic Studies of Active Sites. Blue Copper and Electronic Structural Analogs. Vol. 307. pp. 236–266. doi:10.1021/bk-1986-0307.ch016. ISBN 978-0-8412-0971-8.
{{cite book}}:|journal=ignored (help) - ^ Anderson BF, Baker HM, Dodson EJ, Norris GE, Rumball SV, Waters JM, Baker EN (April 1987). "Structure of human lactoferrin at 3.2-A resolution". Proceedings of the National Academy of Sciences of the United States of America. 84 (7): 1769–73. doi:10.1073/pnas.84.7.1769. PMC 304522. PMID 3470756.
- ^ Wallace, Daniel F (May 2016). "The Regulation of Iron Absorption and Homeostasis". The Clinical Biochemist Reviews. 37 (2): 51–62. ISSN 0159-8090. PMC 5198508. PMID 28303071.
- ^ Rodríguez E, Díaz C (December 1995). "Iron, copper and zinc levels in urine: relationship to various individual factors". Journal of Trace Elements in Medicine and Biology. 9 (4): 200–9. Bibcode:1995JTEMB...9..200R. doi:10.1016/S0946-672X(11)80025-8. PMID 8808191.
- ^ Schümann K, Schäfer SG, Forth W (1986). "Iron absorption and biliary excretion of transferrin in rats". Research in Experimental Medicine. Zeitschrift für die Gesamte Experimentelle Medizin Einschliesslich Experimenteller Chirurgie. 186 (3): 215–9. doi:10.1007/BF01852047. PMID 3738220. S2CID 7925719.
- ^ "Biliary excretion of waste products". Archived from the original on 2017-03-26. Retrieved 2017-03-24.
- ^ Lindskog S (1997). "Structure and mechanism of carbonic anhydrase". Pharmacology & Therapeutics. 74 (1): 1–20. doi:10.1016/S0163-7258(96)00198-2. PMID 9336012.
- ^ Sigel A, Sigel H, Sigel RK, eds. (2008). Metal–carbon bonds in enzymes and cofactors. Metal Ions in Life Sciences. Vol. 6. Wiley. ISBN 978-1-84755-915-9.
- ^ "The Nobel Prize in Chemistry 1964". Nobelprize.org. Retrieved 2008-10-06.
- ^ Hodgkin, D. C. (1965). "The Structure of the Corrin Nucleus from X-ray Analysis". Proc. R. Soc. A. 288 (1414): 294–305. Bibcode:1965RSPSA.288..294H. doi:10.1098/rspa.1965.0219. S2CID 95235740.
- ^ Orme-Johnson, W. H. (1993). Steifel, E. I.; Coucouvannis, D.; Newton, D. C. (eds.). Molybdenum enzymes, cofactors and model systems. Advances in chemistry, Symposium series no. 535. Washington, DC: American Chemical Society. pp. 257. ISBN 9780841227088.
- ^ Chan MK, Kim J, Rees DC (May 1993). "The nitrogenase FeMo-cofactor and P-cluster pair: 2.2 A resolution structures". Science. 260 (5109): 792–4. doi:10.1126/science.8484118. PMID 8484118.
- ^ Packer, L., ed. (2002). Superoxide Dismutase: 349 (Methods in Enzymology). Academic Press. ISBN 978-0-12-182252-1.
- ^ Heinrich P, Löffler G, Petrides PE (2006). Biochemie und Pathobiochemie (in German). Berlin: Springer. p. 123. ISBN 978-3-540-32680-9.
- ^ Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED (June 2004). "Nickel superoxide dismutase structure and mechanism". Biochemistry. 43 (25): 8038–47. doi:10.1021/bi0496081. PMID 15209499.
- ^ Parkin, Alison (2014). "Understanding and Harnessing Hydrogenases, Biological Dihydrogen Catalysts". In Kroneck, Peter M. H.; Sosa Torres, Martha E. (eds.). The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences. Vol. 14. Springer. pp. 99–124. doi:10.1007/978-94-017-9269-1_5. ISBN 978-94-017-9268-4. PMID 25416392.
- ^ Pyle AM (August 1993). "Ribozymes: a distinct class of metalloenzymes". Science. 261 (5122): 709–14. Bibcode:1993Sci...261..709P. doi:10.1126/science.7688142. PMID 7688142.
- ^ Shan S, Yoshida A, Sun S, Piccirilli JA, Herschlag D (October 1999). "Three metal ions at the active site of the Tetrahymena group I ribozyme". Proceedings of the National Academy of Sciences of the United States of America. 96 (22): 12299–304. Bibcode:1999PNAS...9612299S. doi:10.1073/pnas.96.22.12299. PMC 22911. PMID 10535916.
- ^ Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR (August 2015). "New classes of self-cleaving ribozymes revealed by comparative genomics analysis". Nature Chemical Biology. 11 (8): 606–10. doi:10.1038/nchembio.1846. PMC 4509812. PMID 26167874.
- ^ Breaker RR, Joyce GF (December 1994). "A DNA enzyme that cleaves RNA". Chemistry & Biology. 1 (4): 223–9. doi:10.1016/1074-5521(94)90014-0. PMID 9383394.
- ^ Silverman SK (May 2015). "Pursuing DNA catalysts for protein modification". Accounts of Chemical Research. 48 (5): 1369–79. doi:10.1021/acs.accounts.5b00090. PMC 4439366. PMID 25939889.
- ^ Santoro SW, Joyce GF (April 1997). "A general purpose RNA-cleaving DNA enzyme". Proceedings of the National Academy of Sciences of the United States of America. 94 (9): 4262–6. Bibcode:1997PNAS...94.4262S. doi:10.1073/pnas.94.9.4262. PMC 20710. PMID 9113977.
- ^ Breaker RR, Joyce GF (December 1994). "A DNA enzyme that cleaves RNA". Chemistry & Biology. 1 (4): 223–9. doi:10.1016/1074-5521(94)90014-0. PMID 9383394.
- ^ Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y (February 2007). "A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity". Proceedings of the National Academy of Sciences of the United States of America. 104 (7): 2056–61. Bibcode:2007PNAS..104.2056L. doi:10.1073/pnas.0607875104. PMC 1892917. PMID 17284609.
- ^ Torabi SF, Wu P, McGhee CE, Chen L, Hwang K, Zheng N, Cheng J, Lu Y (May 2015). "In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing". Proceedings of the National Academy of Sciences of the United States of America. 112 (19): 5903–8. Bibcode:2015PNAS..112.5903T. doi:10.1073/pnas.1420361112. PMC 4434688. PMID 25918425.
- ^ Stevens FC (August 1983). "Calmodulin: an introduction". Canadian Journal of Biochemistry and Cell Biology. 61 (8): 906–10. doi:10.1139/o83-115. PMID 6313166.
- ^ Chin D, Means AR (August 2000). "Calmodulin: a prototypical calcium sensor". Trends in Cell Biology. 10 (8): 322–8. doi:10.1016/S0962-8924(00)01800-6. PMID 10884684.
- ^ Berg JM (1990). "Zinc finger domains: hypotheses and current knowledge". Annual Review of Biophysics and Biophysical Chemistry. 19 (1): 405–21. doi:10.1146/annurev.bb.19.060190.002201. PMID 2114117.
- ^ Jeoung JH, Fesseler J, Goetzl S, Dobbek H (2014). "Carbon Monoxide. Toxic Gas and Fuel for Anaerobes and Aerobes: Carbon Monoxide Dehydrogenases". In Kroneck PM, Sosa Torres ME (eds.). The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences. Vol. 14. Springer. pp. 37–69. doi:10.1007/978-94-017-9269-1_3. ISBN 978-94-017-9268-4. PMID 25416390.
- ^ Aoki K, Murayama K, Hu NH (2017). "Chapter 7. Solid State Structures of Lead Complexes with Relevance for Biological Systems". In Astrid S, Helmut S, Sigel RK (eds.). Lead: Its Effects on Environment and Health. Metal Ions in Life Sciences. Vol. 17. de Gruyter. pp. 123–200. doi:10.1515/9783110434330-007. ISBN 9783110434330. PMID 28731300.
- ^ Romani, Andrea M. P. (2013). "Magnesium Homeostasis in Mammalian Cells". In Banci, Lucia (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. pp. 69–118. doi:10.1007/978-94-007-5561-1_4. ISBN 978-94-007-5561-1. ISSN 1868-0402. PMID 23595671.
- ^ Roth J, Ponzoni S, Aschner M (2013). "Manganese Homeostasis and Transport". In Banci L (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. pp. 169–201. doi:10.1007/978-94-007-5561-1_6. ISBN 978-94-007-5561-1. ISSN 1868-0402. PMC 6542352. PMID 23595673.
- ^ Dlouhy AC, Outten CE (2013). "The Iron Metallome in Eukaryotic Organisms". In Banci L (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. pp. 241–78. doi:10.1007/978-94-007-5561-1_8. ISBN 978-94-007-5561-1. ISSN 1868-0402. PMC 3924584. PMID 23595675.
- ^ Cracan V, Banerjee R (2013). "Chapter 10 Cobalt and Corrinoid Transport and Biochemistry". In Banci L (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. doi:10.1007/978-94-007-5561-10_10 (inactive 1 November 2024). ISBN 978-94-007-5561-1. ISSN 1868-0402.
{{cite book}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ Sigel A, Sigel H, Sigel RK, eds. (2008). Nickel and Its Surprising Impact in Nature. Metal Ions in Life Sciences. Vol. 2. Wiley. ISBN 978-0-470-01671-8.
- ^ Sydor AM, Zambie DB (2013). "Chapter 11. Nickel Metallomics: General Themes Guiding Nickel Homeostasis". In Banci L (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. doi:10.1007/978-94-007-5561-10_11 (inactive 1 November 2024). ISBN 978-94-007-5561-1. ISSN 1868-0402.
{{cite book}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ Vest KE, Hashemi HF, Cobine PA (2013). "Chapter 13. The Copper Metallome in Eukaryotic Cells". In Banci L (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. doi:10.1007/978-94-007-5561-10_12 (inactive 1 November 2024). ISBN 978-94-007-5561-1. ISSN 1868-0402.
{{cite book}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ Maret W (2013). "Chapter 14 Zinc and the Zinc Proteome". In Banci L (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. doi:10.1007/978-94-007-5561-10_14 (inactive 1 November 2024). ISBN 978-94-007-5561-1. ISSN 1868-0402.
{{cite book}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ Peacock AF, Pecoraro V (2013). "Natural and Artificial Proteins Containing Cadmium". In Sigel A, Sigel H, Sigel RK (eds.). Cadmium: From Toxicity to Essentiality. Metal Ions in Life Sciences. Vol. 11. Springer. pp. 303–337. doi:10.1007/978-94-007-5179-8_10. ISBN 978-94-007-5178-1. PMID 23430777.
- ^ Freisinger EF, Vasac M (2013). "Cadmium in Metallothioneins". In Sigel A, Sigel H, Sigel RK (eds.). Cadmium: From Toxicity to Essentiality. Metal Ions in Life Sciences. Vol. 11. Springer. pp. 339–372. doi:10.1007/978-94-007-5179-8_11. ISBN 978-94-007-5178-1. PMID 23430778.
- ^ Mendel, Ralf R. (2013). "Chapter 15. Metabolism of Molybdenum". In Banci, Lucia (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. doi:10.1007/978-94-007-5561-10_15 (inactive 1 November 2024). ISBN 978-94-007-5561-1. ISSN 1868-0402.
{{cite book}}: CS1 maint: DOI inactive as of November 2024 (link) - ^ ten Brink, Felix (2014). "Living on Acetylene. A Primordial Energy Source". In Kroneck, Peter M. H.; Sosa Torres, Martha E. (eds.). The Metal-Driven Biogeochemistry of Gaseous Compounds in the Environment. Metal Ions in Life Sciences. Vol. 14. Springer. pp. 15–35. doi:10.1007/978-94-017-9269-1_2. ISBN 978-94-017-9268-4. PMID 25416389.
External links
[edit]- Metalloprotein at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Catherine Drennan's Seminar: Snapshots of Metalloproteins
