Galaxy Erasmus MC
The homepage of the Erasmus MC Galaxy community
Galaxy is an open-source platform for FAIR data analysis that enables users to:
- use tools from various domains (that can be plugged into workflows) through its graphical web interface.
- run code in interactive environments (RStudio, Jupyter...) along with other tools or workflows.
- manage data by sharing and publishing results, workflows, and visualizations.
- ensure reproducibility by capturing the necessary information to repeat and understand data analyses.
The Galaxy Community is actively involved in helping the ecosystem improve and sharing scientific discoveries.
News
A new study has sequenced and analyzed two wild loquat chloroplast genomes, revealing key insights into their evolutionary history and the genetic diversity of loquat species.
Hosting R/Shiny applications on Galaxy (and other frameworks)
Thanks to the outcome of a small Galaxy/Bioconductor hackathon earlier this year, it has become much easier to deploy Shiny applications as interactive tools (ITs) on usegalaxy.eu
Opinion: Conda, Anaconda and the much bigger problem behind it
Anaconda, DockerHub, Quay.io, GitHub ... we need to talk about the sustainability of our infrastructure.
Galaxy in Research: Advancing quantum materials research with Galaxy
At Oak Ridge National Laboratory, scientists have harnessed the power of Galaxy to bring together cutting-edge computing resources, paving the way for new discoveries in quantum materials through advanced neutron scattering data analysis.
Galaxy in Research: Exploring DNA methylation and aging across diverse human tissues
How does DNA methylation correlate with aging across different human tissues? A new study provides insights into this question, revealing tissue-specific and shared epigenetic patterns.
Events
Sep 10 - Sep 13A practical introduction to bioinformatics and RNA-seq using Galaxy
Ready to explore the fascinating world of RNA-seq data analysis using Galaxy? Don't miss out on this incredible opportunity! Visit our event webpage now to discover the program details, meet the organising team, and submit your application.
Sep 10 - Sep 12sc-verse Conference 2024
Join us at the sc-verse Conference in Munich to explore the latest advancements in single-cell analysis with Galaxy. Discover the innovative features Galaxy offers in this cutting-edge field of single-cell research, and be sure to attend our poster session for an in-depth discussion and the opportunity to engage directly with our team.
Sep 11Webinar on data management with DataHub (Onedata)
At 14:00 CEST, join this Webinar if you are interested in the recent Galaxy & Onedata integration (remote data source and Object Store).
Oct 7 - Oct 11Galaxy Training Academy 2024
Are you ready to enhance your data analysis skills? The Galaxy Training Academy is a five-day global online event designed for beginners and those looking to enhance their Galaxy expertise.
FTP Upload
For large files, FTP upload is also available on this server. Connect to port 23 with the same credentials you use to log into Galaxy. Once uploaded, open the Galaxy file upload menu, and click on the Choose FTP file button to import the files into your history.
As an example, a screenshot of the configuration settings for FileZilla can be found here.
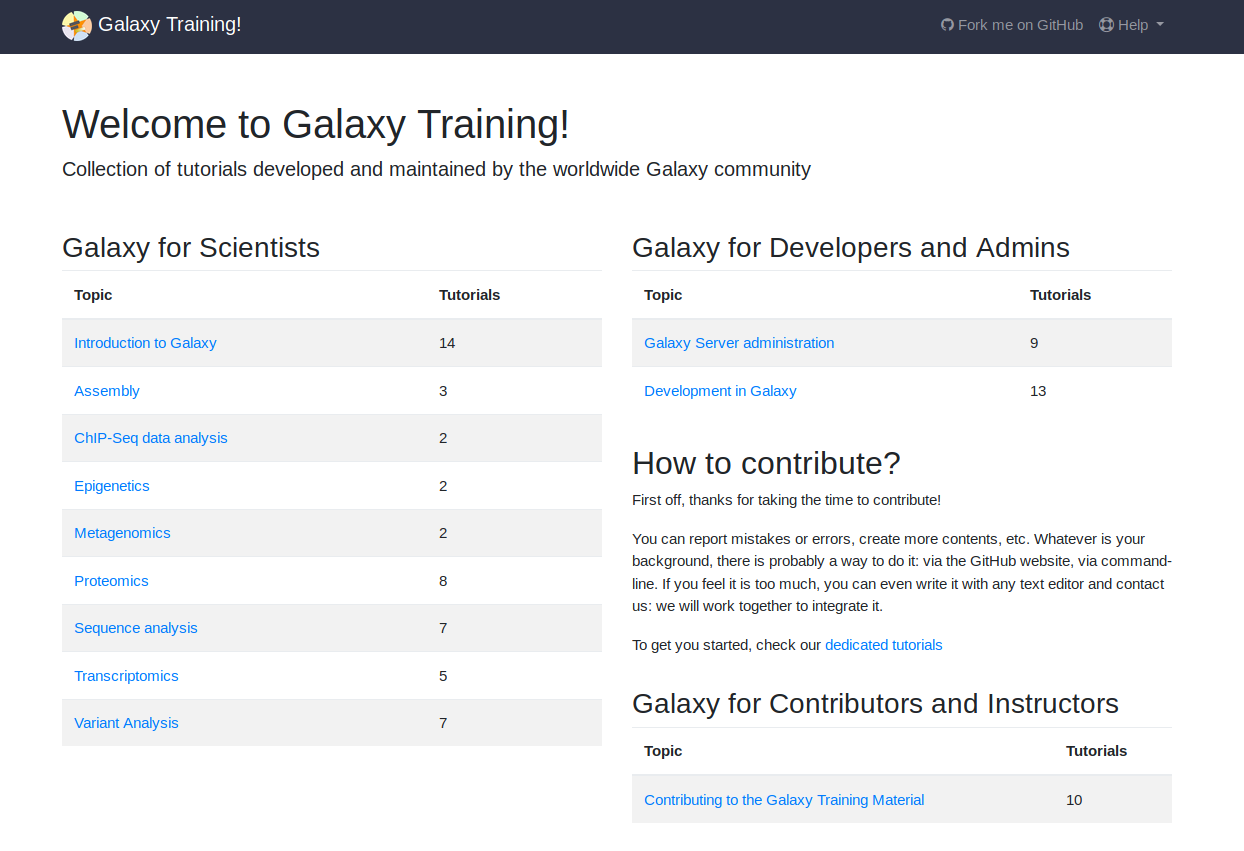
Training
We regularly provide workshops.
But we cannot always meet capacity, so we've put all of our training materials online. This has become a community project with people from all over the world contributing training materials.
Topics include: variant analysis, transcriptomics, metagenomics, epigenetics, and many more!
Our team
This Galaxy is maintained by the Bioinformatics group of the department of Pathology.
For any questions regarding this Galaxy server, please contact us.