Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles
- Jianbin Wang1,
- Benjamin Czech2,
- Amanda Crunk1,
- Adam Wallace1,
- Makedonka Mitreva3,
- Gregory J. Hannon2 and
- Richard E. Davis1,4
- 1Department of Biochemistry and Molecular Genetics, University of Colorado School of Medicine, Aurora, Colorado 80045, USA;
- 2Watson School of Biological Sciences, HHMI, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York 11724, USA;
- 3Genetics and Genome Center, Washington University School of Medicine, St. Louis, Missouri 63108, USA
Abstract
Eukaryotic cells express several classes of small RNAs that regulate gene expression and ensure genome maintenance. Endogenous siRNAs (endo-siRNAs) and Piwi-interacting RNAs (piRNAs) mainly control gene and transposon expression in the germline, while microRNAs (miRNAs) generally function in post-transcriptional gene silencing in both somatic and germline cells. To provide an evolutionary and developmental perspective on small RNA pathways in nematodes, we identified and characterized known and novel small RNA classes through gametogenesis and embryo development in the parasitic nematode Ascaris suum and compared them with known small RNAs of Caenorhabditis elegans. piRNAs, Piwi-clade Argonautes, and other proteins associated with the piRNA pathway have been lost in Ascaris. miRNAs are synthesized immediately after fertilization in utero, before pronuclear fusion, and before the first cleavage of the zygote. This is the earliest expression of small RNAs ever described at a developmental stage long thought to be transcriptionally quiescent. A comparison of the two classes of Ascaris endo-siRNAs, 22G-RNAs and 26G-RNAs, to those in C. elegans, suggests great diversification and plasticity in the use of small RNA pathways during spermatogenesis in different nematodes. Our data reveal conserved characteristics of nematode small RNAs as well as features unique to Ascaris that illustrate significant flexibility in the use of small RNAs pathways, some of which are likely an adaptation to Ascaris' life cycle and parasitism.
Gametogenesis and embryogenesis are developmental periods that see rapid changes in genome organization, gene expression, and cellular identity. Studies of the nematode Caenorhabditis elegans have contributed substantially to our understanding of these processes. Nematodes are present in all environments and are one of the most divergent phyla, with >23,000 described species and an estimated 100,000 to 10 million species (Blaxter 2009). Thus, they offer the opportunity for comparative studies of key developmental processes including small RNA pathways.
Many nematodes are parasitic, and adaptation to the parasitic lifestyle often involves an increase in reproductive capacity and overall body size. The nematode Ascaris is a large (∼25 × 0.5 × 0.5 cm), sexually dimorphic parasite of the vertebrate small intestine. One species infects ∼1 billion people worldwide and is considered an important “neglected disease” (Bethony et al. 2006; Hotez 2008). It is estimated that an Ascaris female has as many as 25,000,000 eggs at one time and lays 200,000 to 2 million fertilized eggs per day (Cram 1925; Brown and Cort 1927; Olsen et al. 1958; Sinniah 1982). Theodor Boveri studied Ascaris at the turn of the 19th Century and first described chromosomes and cytoplasm in heredity, determinate cleavage, and a nematode cell lineage (Satzinger 2008). More recently, Ascaris has been used for a variety of physiological, biochemical, and molecular studies.
Ascaris has several attributes that are advantageous for the analysis of gametogenesis, fertilization, zygote maturation, and early development. First, the long and linear reproductive systems (∼140 cm for males and 240 cm for females) enable the dissection and isolation of discrete stages of gametogenesis as well as fertilized zygotes undergoing a 12- to 24-h maturation in the uterus that precedes pronuclear fusion and development (Fig. 1A). Second, large numbers of synchronized, developmentally staged embryos and larvae (fertilized zygote through the L2 larvae) can be easily obtained (Fig. 1B). Furthermore, a variety of biochemical and molecular tools for studying RNA transcription, processing, translation, and decay have been developed (Hannon et al. 1990a,b; Maroney et al. 1995; Davis et al. 1999; Cohen et al. 2004; Wallace et al. 2010).
Ascaris reproductive systems and early development. (A) Ascaris male and female reproductive systems. The male seminal vesicle contains spermatids that are activated on fertilization of a female. Sperm are stored in the female seminal receptacle. As mature oocytes move through the seminal receptacle, they are fertilized, and for each fertilized egg, an impermeable layer and chitinous shell is immediately formed around the egg in region one of the uterus (zygote-1). The zygote undergoes maturation as it passes through the uterus (within 12–24 h). Note that the figure is not drawn to scale. Extended male and female reproductive systems are ∼140 cm and ∼240 cm in length, respectively. (B) Early Ascaris embryo development. Eggs derived from the region designated as zygote-4 are equivalent to 0-h embryos. Incubation of 0-h embryos at 30°C in high humidity leads to relatively synchronous development (∼85%) as illustrated. The maternal to zygotic transition is thought to occur between 46 and 96 h of development (four to 16 cells). Ascaris larvae L1 and L2 remain within the chitinous shell. The 0-h embryo is ∼100 × 45 μm and does not change in size during development as illustrated. All regions/stages in A and B were analyzed in the study.
Small noncoding RNAs (∼20–30 nucleotides [nt]) have emerged as key players in both transcriptional and post-transcriptional gene regulation; they also play important roles in genome stability and chromatin organization (for reviews, see Ghildiyal and Zamore 2009; Malone and Hannon 2009; Moazed 2009; Bourc'his and Voinnet 2010). While new types of small RNAs are being identified and their classification continues to be revised, a simple system to group small RNAs divides them according to their biogenesis and Argonaute protein binding partners into three major classes: microRNAs (miRNAs), small interfering RNAs (siRNAs), and Piwi-interacting RNAs (piRNAs) (Czech and Hannon 2011). Small RNAs partnered to Argonaute proteins are the core components of the RNA Induced Silencing Complex (RISC). miRNAs are 20–24 nt in size, are derived from genome-encoded hairpins, and function in gene silencing through translational repression or RNA degradation (for reviews, see Bartel 2004, 2009). siRNAs, typically 20–22 nt in size, are often derived from long dsRNA and play a role in heterochromatin formation, silencing of mobile genetic elements, and gene regulation (for reviews, see Okamura and Lai 2008; Carthew and Sontheimer 2009; Ghildiyal and Zamore 2009; Kim et al. 2009). Primary siRNAs in nematodes, fungi, and plants can trigger the generation of secondary siRNAs, a process that involves the action of RNA-dependent RNA polymerases (RdRPs) and serves to amplify the silencing trigger (Ambros et al. 2003b; Chicas et al. 2004; Ruby et al. 2006; Pak and Fire 2007; Sijen et al. 2007). piRNAs are 21- to 30-nt RNAs that specifically associate with Piwi-clade Argonaute proteins and are derived from discrete genomic loci (called piRNA clusters) by a dicer-independent mechanism (for reviews, see Ghildiyal and Zamore 2009; Siomi et al. 2011). piRNAs are present in the germline and are thought to suppress mobile elements and play important roles in spermatogenesis, maintenance of germ cells, and stem cell totipotency (for reviews, see Saito and Siomi 2010; Siomi et al. 2011). Studies in the nematode C. elegans have identified a diversity of small RNA classes, including miRNAs, siRNAs (26G- and 22G-RNAs), and piRNA (21U-RNAs) (Lau et al. 2001; Lee and Ambros 2001; Sijen et al. 2001, 2007; Ambros et al. 2003a; Lim et al. 2003; Sijen and Plasterk 2003; Ruby et al. 2006; Pak and Fire 2007; Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008; Claycomb et al. 2009; Gu et al. 2009; Han et al. 2009; Kato et al. 2009; Stoeckius et al. 2009).
We have profiled Ascaris small RNAs progressively through discrete regions of the reproductive system and stages of early development by deep sequencing. Our data provide an unprecedented window into the evolution and adaptation of small RNAs and their pathways between the free-living C. elegans and a fecund, parasitic nematode that diverged an estimated 400 million years ago (Blaxter 2009). Key findings include remarkable conservation and divergence of nematode miRNAs over 400 million years of evolution. Ascaris endo-siRNAs (26G- and 22G-RNAs), particularly in the testis, demonstrate striking adaptability and plasticity in their expression and biogenesis, including the loss of 3′ 2′-O-methylation of small RNAs, when compared with that of C. elegans. This adaptability may be reflected in part by the loss of piRNAs in Ascaris. Ascaris small RNAs are transcribed and processed during zygote maturation prior to pronuclear fusion in utero, a developmental stage thought to be transcriptionally quiescent (Leatherman and Jongens 2003; Schier 2007; Tadros and Lipshitz 2009). Finally, small RNAs do not target the major eliminated repeat for Ascaris chromatin diminution. Differences in small RNA pathways between Ascaris and C. elegans likely reflect adaptation to the biology and parasitic lifestyle of Ascaris.
Results
Deep sequencing of small RNAs during Ascaris gametogenesis, zygote maturation, and early development
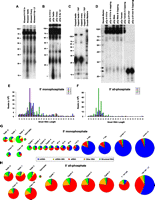
We initially demonstrated the presence of Ascaris small RNAs on polyacrylamide gels by labeling them at either their 5′ or 3′ termini (Fig. 2A,B). We observed two major groups of small RNAs of 21–24 nt and 32 nt in size. Ascaris ∼22-nt RNAs in all stages were readily labeled by capping the RNA with capping enzymes (Fig. 2C), suggesting that some Ascaris small RNAs have 5′ di- or triphosphate termini. To provide a qualitative assessment of the percentage of 5′ di- or triphosphate RNAs, we first 3′ labeled RNAs with 32P-pCp and then capped the RNAs with GTP. A large percentage of the ∼22-nt RNAs increased in size, indicating that the majority of these RNAs could be capped and are 5′ polyphosphate RNAs (Fig. 2D).
Characteristics of Ascaris small RNAs. (A) 5′ end-labeled Ascaris small RNAs. Low-molecular-weight (LMW) enriched RNAs were treated with calf alkaline phosphatase and then 5′ end labeled with 32P using T4 polynucleotide kinase. RNAs in A–D were resolved on 12.5% denaturing PAGE and autoradiographed. (B) 3′ end-labeled Ascaris small RNAs. LMW enriched RNAs were 3′ end-labeled using 32pCp and T4 RNA ligase. (C) Ascaris small RNAs ∼22 nt in length can be cap-labeled. LMW enriched RNAs were capped using 32P-α-GTP and vaccinia guanylyltransferase. Samples were treated with tobacco acid pyrophosphatase (TAP) to remove the cap to confirm cap-labeling. Note the significant labeling of RNAs ∼22 nt in length. (D) The majority of germline and early embryo 22-nt RNAs have 5′ polyphosphates. pCp-labeled small RNAs shown in B were treated with GTP and vaccinia guanylyltransferase. The lower and upper arrows mark the average position of the labeled small RNAs prior to and after capping. Note that the majority of the 32pCp-labeled RNAs are shifted to a longer length. (E,F) Size distribution of analyzed Ascaris small RNAs. (E) All 28 5′ monophosphate libraries characterized combined. (F) All 19 5′ all-phosphate (5′ tri-, di-, and monophosphate) libraries characterized combined. In these and all subsequent size distribution figures, small RNAs starting with A, C, G, and U in different sizes are plotted against their read frequencies (raw reads, before normalization). (G,H) Comparison, classification, and levels of Ascaris small RNAs characterized. (G) 5′ monophosphate RNAs. (H) 5′ all-phosphate libraries derived from different developmental stages. Area of the circles is proportional to the amount of small RNAs present in each stage or tissue after normalization (see text).
Based on these observations, we prepared and analyzed three types of small RNA libraries: (1) libraries of 18- to 34-nt or 18- to 40-nt RNAs with a 5′ monophosphate (5′ monophosphate libraries), (2) libraries of 18- to 34-nt RNAs with a 5′ monophosphate but enriched for 3′ end modifications (5′ monophosphate, 3′ end modified), and (3) libraries of 18- to 28-nt or 18- to 40-nt RNAs with a 5′ tri-, di-, or monophosphate (5′ all-phosphate; summary of these libraries is provided in Supplemental Table S2). These libraries enabled us to identify all expressed small RNAs and characterize the nature of their 5′ and 3′ ends. We failed to detect any significant enrichment of small RNAs in the 5′ monophosphate, 3′ end modified libraries, suggesting that Ascaris small RNAs do not have 3′ modifications (see below). Small RNA libraries were analyzed from different regions of the male and female reproductive systems, spermatids, four stages of zygotes undergoing maturation in the uterus, and staged early embryos from one cell to the larvae to provide an extensive germline and developmental profile (see Fig. 1; Supplemental Table S2). To enable our analysis of the small RNA libraries, we also conducted genome and transcriptome sequencing (for an overview of these data, see Table S1). These data helped us to define different types of small RNAs, their biogenesis, targets, and potential function.
We initiated our analysis by plotting all reads that perfectly matched the genome or transcriptome (J Wang, M Mitreva, M Blaxter, M Berriman, RE Davis, in prep.) as a function of length (Fig. 2E,F; Supplemental Figs. S1–S4). The vast majority of Ascaris small RNAs are 21–24 nt in length with a less abundant fraction that is 26 nt in length (Fig. 2E,F; Supplemental Figs. S1–S4). Annotation of our small RNA sequences indicated that the small RNAs 21–24 nt in length are primarily miRNAs and endo-siRNAs, whereas the 26-nt reads show a strong bias to start with guanosine. Thus, in analogy to 26G-RNAs in C. elegans, we refer to these small RNAs as 26G-RNAs. We also identified larger RNAs that originate from atypical hairpins (see below). Remaining reads matching abundant cellular RNAs were summarized as “structural RNAs” (snRNAs, snoRNAs, rRNAs, tRNAs, etc.), which made up ∼10% of all reads (Fig. 2G,H; Supplemental Table S2). When the small RNA reads between libraries from different stages were compared, we observed that the total number of small RNAs increases progressively (per amount of total RNA/sample), particularly from 128-cell embryos through the larval stages. We found that endo-siRNAs are the predominant small RNA class in the germline. In addition, our data demonstrate a transition in the major type of small RNAs from endo-siRNAs to miRNAs during larval organogenesis and morphogenesis (Fig. 2G,H), as suggested by earlier reports in C. elegans (Martinez et al. 2008; Stoeckius et al. 2009). Our data, however, for the first time clearly define the temporal dynamics and identities of these RNA changes.
Lack of piRNA pathway components and small RNAs in Ascaris
Piwi-clade proteins and piRNAs have been found in metazoa ranging from animal phyla that diverged before the emergence of the Bilateria (sponges and cnidarians) through worms, flies, and humans (Grimson et al. 2008). piRNAs are derived from distinct genomic loci and are methylated on their 3′ terminal 2′ oxygen by HEN1 (Ghildiyal and Zamore 2009; Siomi et al. 2011). piRNA orthologs in C. elegans are only 21 nt in length, have a 5′ terminal U, bind Piwi-clade Argonautes, and are known as 21U-RNAs. These piRNAs are not conserved at the sequence level among Caenorhabditis species and do not exhibit complementarity to targets. However, they share a common upstream motif in their genomic loci (Ruby et al. 2006; Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008; de Wit et al. 2009; Kato et al. 2009).
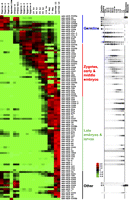
Our data provide several lines of evidence that the piRNA pathway has been lost in Ascaris. First, our direct sequencing of discrete regions of both the male and female germlines did not reveal any small RNAs with characteristics of piRNAs (Fig. 3A,B; Supplemental Figs. S1–S4; Supplemental Material on 21U/piRNA). Second, we were not able to identify Piwi-clade Argonaute orthologs using bioinformatics on our extensive genomic (18× coverage of the genome) and cDNA sequence data from testis and ovary tissue (Supplemental Table S1). However, we were able to identify cDNA sequences encoding 10 other Argonautes proteins of the AGO and WAGO clades (Fig. 3C) as well as a variety of proteins associated with small RNA biogenesis and function (Supplemental Table S9). Third, we also failed to detect an ortholog of the HEN1 methyltransferase in the Ascaris genome or germline transcriptome. In addition, no small RNAs were identified with 3′ methyl modifications (Fig. 3D–H). Notably, Piwi Argonaute and Hen1 orthologs are also not identifiable in a related parasite, Brugia malayi (Ghedin et al. 2007). Given that piRNAs have been shown to be present in metazoa that emerged even before the first bilateral animals (Grimson et al. 2008), the most parsimonious explanation for the absence of piRNAs in Ascaris is that these small RNAs and their associated proteins, Piwi and Hen1, have been lost.
Ascaris lacks the piRNA pathway. (A,B) No piRNAs are present in Ascaris, as illustrated by the size distribution of Ascaris germline small RNAs. (A) Testis small RNAs. (B) Ovary small RNAs. Raw reads (before normalization) from the libraries are plotted. Note that no 21U or other U-initiated small RNAs are enriched in either the testis or ovary after miRNAs and siRNAs are removed. This is also observed for other germline and embryo libraries (see Supplemental Data). (C) Ascaris lacks Piwi-clade Argonaute protein. ClustalX was used to generate an alignment of Ascaris and C. elegans Argonaute proteins. The alignment was used to generate a distance matrix and then analyzed by neighbor-joining and bootstrapping methods using PHYLIP software. TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used to display the resulted tree. Numbers on the branches show the support value for 1000 bootstraps. Argonaute proteins from Ascaris are colored in blue and red for AGO-clade and WAGO-clade proteins, respectively. C. elegans Piwi-clade Argonautes are colored in green. Accession numbers for the Ascaris proteins can be found in Supplemental Table S9. Note that Ascaris also lacks an ortholog of C. elegans RDE-1. (D) RNAs with a 2′-O-methyl are resistant to periodate treatment and β-elimination. Control RNAs (22 nt) were 5′ labeled by kinasing and then either mock treated (−) or treated with periodate followed by β-elimination (+).(E) Ascaris 26G-RNAs are not resistant to periodate/β-elimination and thus are not 3′ modified. Northern blots of Ascaris RNAs treated with periodate/β-elimination and probed with Ascaris 26G-1 and 26G-4 probes. (F) Ascaris 22G-RNAs are not 3′ modified. Northern blot as in E using an Ascaris 22G-RNA probe. (G) Ascaris miRNAs are not 3′ modified. Northern blot as in E using an Ascaris miRNA probe (asu-miR-5342). (H) C. elegans 21U RNAs are 3′ modified and resistant to periodate/β-elimination. Northern blot of C. elegans RNAs treated with periodate/β-elimination and probed with a C. elegans 21U-RNA probe.
Ascaris miRNAs conservation and expression
We next used de novo analyses to identify miRNAs as described in the Supplemental Material. We identified 97 miRNAs that were grouped into 59 Ascaris seed families (Fig. 4; Supplemental Fig. S5; Supplemental Tables S3–S5). The seed sequences for 78 (80%) Ascaris miRNAs are conserved in other eukaryotic miRNAs (Supplemental Table S4). When considering the miRNAs and their hairpin conservation on the sequence level (see Supplemental Methods), 64 (66%) of the Ascaris miRNAs are conserved in other metazoa (Supplemental Table S5). With respect to nematodes, the majority of known miRNAs in the closely related parasitic nematode B. malayi were found in Ascaris (Supplemental Table S5). However, to date only ∼30 B. malayi miRNAs are known; many of these were defined only by computational methods and sequence conservation, and thus they likely represent the most conserved miRNAs (Poole et al. 2010). For more distantly related nematodes, about 30% and ∼24% of known miRNAs Caenorhabditis and Pristionchus are conserved in Ascaris, respectively (Supplemental Table S5). A high percentage of conserved miRNAs among closely related nematodes has also been observed within the Caenorhabditis group (>90%), and between Caenorhabditis and Pristionchus (∼70%) (de Wit et al. 2009). When Ascaris miRNAs are compared with those of fruit flies and humans, conservation drops to ∼18% and ∼5%, respectively. Interestingly, of the 33 Ascaris-specific miRNAs, 16 are derived from polycistronic loci, and these miRNAs are primarily expressed in the germline, zygote maturation, and early embryos (Fig. 4; Supplemental Fig. S7; Supplemental Tables S3–S5).
Dynamic miRNA expression profiles in Ascaris development. Heatmap and Northern Blot expression profiles of Ascaris miRNAs. For each miRNA, colors represent log2 values of fold changes to the average read frequencies of the miRNA during development. Heatmap expression profiles represent normalized mature miRNA read numbers.
Ascaris miRNAs are dynamically expressed during development. The range of Ascaris miRNA abundances in individual developmental stages is very dynamic, spanning six orders of magnitude. To validate the small RNA abundance as determined by the small RNA reads in the libraries, we performed Northern blotting for 42 individual miRNAs (43%) covering different expression patterns (Fig. 4). We found a very strong correlation between Northern blot data and read frequencies, with a mean and median Pearson correlation coefficient r-value of 0.85 and 0.91, respectively (Supplemental Table S3), demonstrating that our sequencing profiles are quantitative. Based on their expression patterns, Ascaris miRNAs were divided into four groups: (1) germline expressed miRNAs; (2) miRNAs predominating in the zygote, early, and middle embryo stages; (3) late embryo and larval expressed miRNAs; and (4) other miRNAs (Fig. 4). Of these 97 miRNAs, none were highly expressed throughout development. This argues against the idea of “housekeeping” miRNAs necessary for basic cellular functions. Several miRNAs are highly expressed during early development and then lost over a narrow developmental window (two- to 128-cell embryos). These miRNAs are often derived from polycistronic loci with similar seeds, suggesting coordinated target regulation (Supplemental Fig. S7). Information on the features of Ascaris miRNAs such as miRNA/miRNA* sequences and frequencies, hairpin sequences, polycistronic clusters, expression profiles, RNA editing, nontemplated 3′ end modifications, as well as comparison of seed families and miRNAs with other organisms is provided in Supplemental Material. Overall, our data suggest that while there is high conservation of some miRNAs within nematode lineages, there is also significant miRNA evolution and diversity.
Early expression of Ascaris miRNAs during zygote maturation
Surprisingly, at least 15 miRNAs appear immediately after fertilization, before pronuclear fusion, and during maturation of the zygote (Figs. 4, 5A; Supplemental Fig. S6). Note that immediately following fertilization, the zygote synthesizes a chitinous shell followed by an impermeable layer that surrounds the zygote. Northern blot analysis for these new zygotic miRNAs did not identify any mature miRNA or miRNA precursors in Ascaris spermatids or oocytes, nor did we see accumulations of primary or pre-miRNAs in the early zygotes, suggesting that these miRNAs are likely derived from new transcription (see Supplemental Fig. S6A). These miRNAs increase in abundance as the zygote matures in the uterus. Other miRNAs also appear or increase in frequency as early as the one- to two-cell stage, and they increase as the embryo develops (Fig. 4; Supplemental Table S3). A recent study on early C. elegans embryos (Wu et al. 2010) described a miRNA seed family whose miRNAs (miR-35-42) were of maternal origin. Our data show the corresponding seed family in Ascaris (36a/36b/36c/36d/36e/36f/5348) is of embryonic origin.
Ascaris endo-siRNAs in gametogenesis and embryogenesis
We identified a large number of dynamically expressed small RNAs that are not miRNAs, do not appear to be derived from hairpin structures, and are not related to structural RNAs (rRNA, tRNA, snRNAs, etc.). We have defined these Ascaris small RNAs as endo-siRNAs. These RNAs are enriched in the germline and early embryos of Ascaris (Fig. 2G,H; Supplemental Figs. S1–S4). Most of these small RNAs start with G and are either 22 nt or 26 nt in length, reminiscent of C. elegans 22G- and 26G-RNAs. C. elegans 22G-RNAs function in chromosome segregation, transposon silencing, and gene regulation (Ambros et al. 2003b; Lim et al. 2003; Ruby et al. 2006; Pak and Fire 2007; Sijen et al. 2007; Claycomb et al. 2009; Gent et al. 2009; Gu et al. 2009; van Wolfswinkel et al. 2009; Vasale et al. 2010; Zhang et al. 2011) C. elegans has two classes of 26G-RNAs that regulate gene expression associated with spermatogenesis (Class I) and early zygotic development (Class II) (Han et al. 2009; Conine et al. 2010; Gent et al. 2010; Vasale et al. 2010; Zhang et al. 2011).
A subset of Ascaris endo-siRNAs correspond to repetitive elements, such as DNA transposons and retrotransposons, and are expressed at relatively high levels as has been observed in C. elegans (Supplemental Table S8; Gu et al. 2009). However, most of the Ascaris endo-siRNAs are not highly expressed and correspond either to unique genome sequences or to a broad spectrum of different mRNA transcripts (Fig. 6). For the mRNA-matching endo-siRNAs, the majority are in the antisense orientation (Fig. 6A,B). Most of these RNAs start with G and are of two distinct lengths, 22 nt and 26 nt (Fig. 6A). Based on the orientation of the endo-siRNAs to mRNAs, their length, and starting bases, we grouped them into the four classes: (1) As.22G, 21- to 24-nt small RNAs beginning with a G and antisense to mRNAs; (2) As.26G, 25- to 27-nt small RNAs beginning with G and antisense to mRNAs; (3) As.22H, other small RNAs antisense to mRNAs that do not begin with G; and (4) Sense, small RNAs sense to mRNAs. A comparison between 17 pairs of 5′ monophosphate and 5′ all-phosphate libraries definitively demonstrates that As.22G and As.22H are highly enriched in the 5′ all-phosphate libraries, in contrast to As.26G and Sense RNA that are not enriched (Fig. 6B,C,E). The percentage and enrichment of 22-nt RNAs (As.22G and As.22H) in the 5′ all-phosphate libraries is consistent with most ∼22-nt RNAs in Ascaris having 5′ termini with di- or triphosphates (Fig. 2D). Enzymatic treatment of RNA followed by Northern blot analyses further demonstrated that Ascaris 22-nt RNAs have 5′ polyphosphates (Fig. 5D–F).
Ascaris small RNAs expression and their 5′ phosphate termini. (A) Northern blots illustrating intrauterine zygotic expression of miRNAs (for whole blots, see Supplemental Fig. S6A). (B) Northern blots illustrating dynamic expression of 22G-RNAs. (C) Northern blots illustrating expression of testis-specific 26G-RNAs. (D) Ascaris miRNAs have a 5′ monophosphate. Northern blot of Ascaris LMW RNA untreated or treated with terminator, GTP and guanylyltransferase (capping), or ligation of an RNA oligonucleotide (T4 RNA ligation) and probed with asu-miR-5347 probe. Note that the miRNA is degraded by terminator, is uncapped, and can be ligated to an RNA oligonucleotide, indicating the 5′ end is a monophosphate. (E) Ascaris 22G-RNAs have 5′ polyphosphates. Northern blot as in D probed with a 22G-RNA probe. Note that the 22G-RNA can be capped but is not susceptible to terminator or T4 RNA ligation, suggesting the 5′ end of the RNA is a di- or triphosphate. (F) Ascaris 26G-RNAs have a 5′ monophosphate. Northern blot as in D probed with a 26G-RNA probe. Note that the 26G-RNA is testis specific and is degraded by terminator, is uncapped, and can be ligated to an RNA oligonucleotide, indicating the 5′ end is a monophosphate. The band at ∼32 nt in the RNA+T4 RNA ligation is an artifact in the ovary.
Ascaris endo-siRNAs to mRNAs. (A) Size distribution of small RNA raw reads corresponding to mRNAs from 17 5′ all-phosphate libraries. (Upper panel) Antisense small RNAs; (lower panel) sense small RNAs. (B) Comparison, classification, and levels of normalized small RNAs corresponding to mRNAs. Area of the circles is proportional to the amount of different types of small RNAs (see text) present in the 5′-mono- and 5′-all-phosphate libraries. (C) Twenty-two-nucleotide small RNAs are polyphosphate RNAs, whereas 26G-RNAs are monophosphate RNAs. miRNAs were used to normalize the reads between 5′ monophosphate and 5′ all-phosphate libraries. In these and all subsequent box-and-whisker plots, the top and bottom ends of each box represent the 75th and 25th percentile, respectively; the middle line represents the median value; the extended lines illustrate the range (highest and lowest value); and dots represent values beyond the extremes of the whisker. (D) siRNA distribution on mRNA transcripts. Different types of small RNA reads from 5′ all-phosphate libraries are plotted for 8279 full-length cDNAs. Each mRNA is equally divided into 20 bins, and small RNAs mapped to each bin were counted. Plotted is the sum of the counts (y-axis) against the relative position (x-axis) on the cDNA from 5′ to 3′, with the 5′ end of the mRNA designated as 1 and the site of polyadenylation at the 3′ end designated as 100. (E) Twenty-two-nucleotide monophosphate RNAs are likely derived from 22-nt polyphosphate RNAs. Plots illustrate a comparison of small RNA reads between 5′ all-phosphate libraries (x-axis) and 5′ monophosphate libraries (y-axis). The data sets are the top 200 siRNAs in read frequency.
Ascaris As.26G RNAs are specifically expressed in testis (Figs. 2G,H and 5C,F). These RNAs have features similar to C. elegans Class I 26G-RNAs that are involved in spermatogenesis (Ruby et al. 2006; Han et al. 2009; Conine et al. 2010) and are believed to be their orthologs: Both of them are 26 nt in length, start with G, and have 5′ monophosphates (Fig. 5F); both are antisense to testis-enriched mRNAs associated with spermatogenesis (see below). Ascaris also has an orthologous testis-specific Argonaute protein to C. elegans testis-specific Argonautes ALG-3/4 that are associated with Class I 26G-RNAs (Fig. 2C; Supplemental Table S9). Ascaris appears to lack the C. elegans early zygotic development (Class II) 26G-RNAs (Fig. 2G,H; Vasale et al. 2010). Interestingly, unlike C. elegans 26G-RNAs, Ascaris 26G-RNAs are not 3′ modified as shown in β-elimination analyses (Fig. 3D–F).
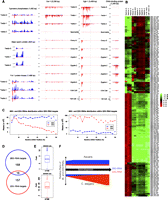
The distribution of endo-siRNAs on their corresponding mRNA targets differs between Ascaris and C. elegans. 22G-RNAs in C. elegans show enrichment at both mRNA termini, and it has been suggested that the initiating events for 22G synthesis originate at the 3′ end of the mRNA template followed by progressive 5′ cycles of transcription by RdRP (Gu et al. 2009). In contrast, Ascaris 22G-RNAs and 22H-RNAs are distributed across their mRNA targets increasing in frequency toward the 5′ end with an abrupt drop-off at the boundary between the 5′ untranslated region (UTR) and open reading frame (ORF) (Figs. 6D, 7A,C). This may indicate a 5′ bias in the recruitment of RdRP complexes in Ascaris. Alternatively, initial formation of small RNAs at the 3′ end of the mRNA could lead to progressively increased 5′ recruitment of RdRP complexes. The observed differences in the distribution of 22G-RNAs on their target mRNAs suggest significant differences in the biogenesis of 22G-RNAs between Ascaris and C. elegans. Interestingly, Ascaris 26G-RNAs show a distinct pattern with the small RNAs mainly associated with ORFs (Figs. 6D, 7A,C). The primary distribution of Ascaris 26G-RNAs on mRNA ORFs leads us to speculate that the regions of the mRNA targeted (and perhaps what mRNAs get targeted) may be associated with some aspect of translation or the location of 80S ribosomes on mRNAs.
Ascaris 26G-RNAs and 22G-RNAs targets. (A) Examples of distribution of 26G- and 22G-RNAs to mRNAs throughout development. Note that all small RNAs are antisense. (Red) 22G-RNAs; (blue) 26G-RNAs. Vertical dashed lines indicate the start and stop of the ORF. Numbers on left indicate the normalized read frequency of the gray line. Note that for 26G-RNA targets, only the profiles from testis 26G-RNAs are shown; for their complete profiles, see Supplemental Table S7 and Supplemental Figures S9 and S10. (B) Dynamic expression profiles of 22G-RNAs to 159 exemplary targets (see text). Normalized As.22G RNA reads mapping to each target mRNA were used for the heatmap. For each target, colors represent log2 values of fold changes to the average read frequencies of the As.22G RNAs during development. (C) Distribution of 26G- and 22G-RNAs on mRNAs predominantly targeted by one of the two types of small RNAs. Only mRNAs identified as near full length are used and include 138 26G-targeted mRNAs and 97 22G-targeted mRNAs (see Supplemental Table S7; Supplemental Figs. S9, S10). Plotted is the sum of the counts (y-axis) against the relative position (x-axis) on the cDNA from 5′ to 3′, with the 5′ end of the mRNA designated as 1 and the site of polyadenylation at the 3′ end designated as 100. (D) Venn diagram illustrating the relationship between the mRNAs targeted by 26G- or 22G-RNAs. (E) Ratio of defined testis 26G-RNA/22G-RNAs mRNA targets. (Upper panel) 26G-RNA targets, the 26G/22G ratio; (lower panel) 22G-RNA targets. (F) Expression profile of 26G-RNAs and 22G-RNAs in spermatogenesis is different for Ascaris and C. elegans (see Discussion). Note that for C. elegans, the total level of 26G-RNAs decreases during spermatogenesis with a concomitant and dramatic increase in 22G-RNAs targeting the same mRNAs. In contrast, the ratio of Ascaris 26G/22G RNAs to mRNAs remains the same during spermatogenesis and the 26G-RNAs are always the dominant small RNA species. Note that the figure does not represent absolute levels of the RNAs.
Another type of 22-nt RNAs in Ascaris match the genome but not our comprehensive transcriptome data. These small RNAs most likely correspond to nonpolyadenylated transcripts, RNA intermediates, or nonexpressed loci as observed in C. elegans (see other RNA in Fig.2G,H).
We observed small RNAs to the sense strand of mRNAs that do not show a preference for the first base and have a broader size distribution up to 40 nt (Fig. 6A). This resembles the distribution of degraded structural RNAs (Supplemental Figs. S1–S4), suggesting they might be mRNA degradation products. However, we also observed a small peak of RNAs 21–23 nt in length for the sense small RNAs, suggesting these small RNAs could potentially be another functional small RNA class (Fig. 6A).
Ascaris 26G-/22G-RNA targets and small RNA diversification in spermatogenesis
To gain insights into the biological roles of Ascaris 26G- and 22G-RNAs, we defined a set of exemplary small RNA targeted mRNAs using a cutoff of a minimum of 100 reads/mRNA/million reads for 26G-RNAs or 22G-RNAs in any of the 5′ all-phosphate library (Fig. 7; Supplemental Figs. S9, S10; Supplemental Table S7). This generated 184 and 173 mRNAs for the 26G- and 22G-RNA targets, respectively, with an overlap of 16 targets (Fig. 7D). We refined these targets based on the 26G/22G ratio (see Supplemental Methods), and 182 and 159 exemplary targets for 26G- and 22G-RNAs were presented (Fig. 7; Supplemental Figs. S9, S10; Supplemental Table S7). The 182 exemplary mRNAs targeted by 26G-RNAs mainly encode three types of proteins: (1) protein kinases (51), (2) protein phosphatases (24), and (3) major sperm proteins (18). These proteins are known to be associated with C. elegans spermatogenesis (Reinke et al. 2000). A comparison of RNA-seq reads from Ascaris testis, ovary, embryo, and larvae showed that the 182 mRNAs targeted by 26G-RNAs are highly enriched in the testis (Supplemental Table S7). In contrast, the 159 exemplary mRNA targeted by 22G-RNAs encode a functionally diverse population of proteins important in development and are more abundant in the Ascaris ovary (Supplemental Table S7). More than a dozen of these mRNAs encode DNA/chromatin binding proteins, such as transcription factors MEF-2, NHR-25, and FZR-1, as well as histone H3 and H4. The expression of the 22G-RNAs is highly dynamic (Fig. 7B). Their expression profiles appear to match their potential biological roles: 22G-RNAs increase dramatically during and after eggshell formation occurs against the mRNA encoding a chitin-binding protein (ASCF_6433_1340) (Fig. 7A); 22G-RNAs are specifically expressed in the germinal regions of the ovary against histone mRNAs (ASCF_5548_1533 and ASCF_7825_1030) (Supplemental Fig. S9); and 22G-RNAs are highly expressed in the testis against the CGH-1 mRNA, which encodes a protein involved in mRNA regulation (ASCF_1262_3459) (Fig. 7A). Interestingly for CGH-1, not only the level but also the distributions of 22G-RNAs on the mRNA are developmentally regulated.
Ascaris ∼28-nt RNAs derived from atypical hairpins
5′ and 3′ labeling of Ascaris small RNAs revealed RNAs of ∼32 nt with 5′ monophosphates (Fig. 2A,B). Many of these small RNAs are derived from snoRNAs and tRNAs, but a small subset of ∼28-nt RNAs in 5′ monophosphate libraries exhibited differential expression patterns and are derived from atypical genomic hairpins (Supplemental Figs. S11, 12). These small RNAs show 5′ end homogeneity with a corresponding lower abundance RNA from the other side of the hairpin. Several are derived from families (Up5 has five members, and Up2 has two members). These small RNAs are more abundant in the zygote and one-cell embryos (0 and 24 h) (Supplemental Fig. S11B). Northern blot analysis for these small RNAs indicated size heterogeneity (28–32 nt in length) and putative, more abundant precursor transcripts of 70–80 nt (Supplemental Fig. S11C). These precursor transcripts do not correspond to snoRNA sequences or their derivatives, and they are not conserved in other studied nematodes. The enrichment of several of these RNAs in the zygote and early embryos suggests developmentally regulated expression. A low ratio of small RNA to a larger precursor RNA was previously observed for snoRNA or tRNA-derived small RNA, several of which were shown to be involved in RNA silencing (Ender et al. 2008; Taft et al. 2009; Haussecker et al. 2010). Thus, these small RNAs potentially represent a novel small silencing RNA class in Ascaris.
Discussion
This is the first study in nematodes to systematically catalog the expression of small RNAs through regions of the male and female reproductive systems, during zygote maturation, and all stages of early development. We characterized the differential expression profiles of miRNAs, 22G endo-siRNAs, 26G endo-siRNAs, other endo-siRNAs, and larger RNAs derived from atypical hairpins. Few comparative small RNA analyses have been carried out on nematodes, particularly from distant phylogenetic species (de Wit et al. 2009; Poole et al. 2010). Our comparative analyses demonstrate significant conservation of some miRNAs and endogenous siRNA classes between Ascaris suum and C. elegans. However, there is also considerable diversity in miRNAs as well as adaptation of endo-siRNA biogenesis and their targets. In particular, piRNAs and protein components of the piRNA pathway have been lost in Ascaris. Furthermore, endo-siRNAs involved in Ascaris spermatogenesis have increased and diverged as compared with C. elegans, perhaps to complement the functional consequences of piRNA loss.
Ascaris miRNAs and endo-siRNAs are differentially expressed during gametogenesis and throughout development to the L2 larvae. Endo-siRNAs are the dominant class of small RNAs in the male and female germline and through the 10- to 26-cell stage of development. Thereafter, the number and abundance of specific miRNAs increase and predominate in the larvae, while the levels of endo-siRNAs significantly decrease (Fig. 2G,H). This transition indicates that siRNA pathways are dominant in the germline, whereas miRNA pathways prevail in embryo and larval development. This transition is likely to be the case in C. elegans (Martinez et al. 2008; Stoeckius et al. 2009), but no high-resolution staged samples or quantitative measurements have been used to indicate the exact developmental time and scale of this transition.
Expression of small RNAs prior to pronuclear fusion during zygotic maturation
A key and unexpected finding is that small RNAs are synthesized in the zygote within the uterus before pronuclear fusion. Following fertilization in most metazoa, new zygotic transcription does not occur until the maternal to zygotic transition, which in nematodes begins at the four- to eight-cell stage (Cleavinger et al. 1989; Edgar et al. 1994; Baugh et al. 2003). Previous studies in Ascaris indicated that maternal rRNAs are not contributed to the oocyte as observed in most metazoa, but that rRNAs for early development are newly transcribed in the male pronucleus immediately following fertilization (Kaulenas and Fairbairn 1968). The Ascaris zygote undergoes maturation during the 12- to 24-h it passes through the uterus; this maturation is necessary for subsequent early development (Fairbairn 1957). A study from Muller's group demonstrated that mRNA derived from the FERT-1 gene are transcribed during intrauterine zygote maturation (Spicher et al. 1994). It remains to be determined whether the miRNAs synthesized prior to pronuclear fusion are derived from the male or female pronucleus or both. Thus, gene expression in post-fertilization development in Ascaris exhibits a number of novel features including miRNA transcription occurring during zygote maturation, earlier than in any system yet described and at a developmental stage long thought to be quiescent. When the mature eggs (zygote-4 or 0 h) are released from the Ascaris uterus and then passed from the mammalian host to the external environment, development begins with pronuclear fusion (Fig. 1), and additional new miRNAs appear at the one- to two-cell stage of development (Figs. 4, 5A). Our data suggest that RNA pol II transcription starts earlier in embryogenesis than previously thought for either Ascaris or C. elegans (Cleavinger et al. 1989; Edgar et al. 1994; Baugh et al. 2003).
Role of small RNAs in early embryo development
Notably, Ascaris 22G endo-siRNAs are also synthesized in the zygote prior to pronuclear fusion and get contributed to early embryos. Studies in C. elegans demonstrated that dicer and miRNA-associated ALG-1/2 Argonaute mutants led to discrete developmental phenotypes. However, recent studies in mice suggest that early developmental phenotypes of dicer mutants may not be directly related to the loss or reduction of miRNAs but to the alteration of endogenous siRNA pathways (Ma et al. 2010; Suh et al. 2010). Endo-siRNAs that are maternally contributed or differentially expressed during early development may play a role in the maternal to zygotic transition and other important roles in early development, including the onset of primordial germ cells. Additional studies are needed to clarify the roles of these endogenous siRNAs during early development.
Elimination of individual miRNAs or miRNA seed families in C. elegans leads to developmental phenotypes including death of embryos or early larvae and defects in locomotion, body size, egg laying, and dauer larvae formation (Miska et al. 2007; Alvarez-Saavedra and Horvitz 2010; Brenner et al. 2010; Shaw et al. 2010). Multiple Ascaris miRNAs expressed in early development also belong to the same seed families and share similar expression patterns (Supplemental Table S4), such as Ascaris seed family miR-36a/36b/36c/36d/36e/36f/5348 (C. elegans miR-35/36/37/38/39/40/41/42), miR-791/5350a/5350b/5350c/5350d/5351/5367 (C. elegans miR-790/791), miR-44a/279a/279b/279c (C. elegans miR-44/45/61/247), and miR-2a/2b/43a/43b/43c/43d/43e/250 (C. elegans miR-2/43/250/797). Our studies clearly demonstrate the timing for the specific developmental expression of these miRNAs and their conserved seed families (Fig. 4; Supplemental Tables S3, S4). In several cases, miRNAs in the same seed families are expressed in a narrow window during embryogenesis. Redundant seed families could ensure silencing at key points in development and/or expression of appropriate seed miRNAs in different cells during early development. It will be interesting to determine the targets for these miRNA in Ascaris and compare them with those in C. elegans.
We observed a dozen miRNAs maternally contributed to the zygote, but no clear example of paternal miRNA contributions. In C. elegans, maternally contributed and some zygotic miRNAs (Kato et al. 2009; Stoeckius et al. 2009; Wu et al. 2010) appear to play important roles in early development (Miska et al. 2007; Alvarez-Saavedra and Horvitz 2010; Brenner et al. 2010; Shaw et al. 2010) as they do in mice, Xenopus, and flies (Bernstein et al. 2003; Leaman et al. 2005; Martello et al. 2007; Tang et al. 2007). miRNAs are also known to contribute to the maternal to zygotic transition in zebrafish and flies, although the miRNAs involved are not conserved (Giraldez et al. 2006). Ascaris 22G-RNAs are also maternally contributed, synthesized in the zygote prior to nuclear fusion, and differentially expressed through the 24-cell stage of embryos. The role of germline contributed small RNAs, early zygotic transcription of miRNAs in Ascaris and C. elegans, and new endo-siRNAs during early development remain to be determined. Interestingly, the majority of the miRNAs unique to Ascaris are derived from the germline, zygote, and early stages of development, whereas those Ascaris miRNAs conserved in C. elegans are expressed in later stages of development (26 cell–L2). We hypothesize that the Ascaris-specific miRNAs are related to Ascaris sexual dimorphism and differences associated with Ascaris development and metabolism.
Small RNAs in Ascaris spermatogenesis
Ascaris has lost the piRNA pathway. Considering the important role of piRNAs in spermatogenesis and silencing mobile elements in the germline with the exceptional reproductive capacity in Ascaris (see above), other small RNAs or other mechanisms must serve this key function in Ascaris. Consistent with this hypothesis, Ascaris 26G- and 22G-RNAs are the dominant Ascaris testis small RNAs. Based on analysis of the biogenesis of C. elegans 22G-RNAs in dicer mutants, Gu et al. (2009) proposed that 22G-RNAs are generated through a dicer (DCR-1) independent pathway. They further noted that the WAGO 22G-RNA system in C. elegans is similar in many respects to Drosophila and vertebrate piRNA pathways. In addition, piRNAs in C. elegans have been demonstrated to act upstream of 22G-RNA biogenesis (Batista et al. 2008; Das et al. 2008). Thus, Ascaris 26G- and 22G-RNAs may have adapted to serve additional roles typically played by C. elegans piRNAs. Functional overlap of piRNA, 26G-, and 22G-RNAs, including targeting mobile elements, likely occurs in C. elegans as temperature-sensitive spermatogenesis defects resulting from Piwi-related PRG-1 mutants are similar to those observed for 26G- and 22G-RNA defects (Batista et al. 2008; Wang and Reinke 2008; Conine et al. 2010).
Spermatogenesis is a very temperature-sensitive process in C. elegans (L'Hernault 2009). Many of the mutations in the male germline and spermatogenesis cause temperature-sensitive phenotypes (Batista et al. 2008; Das et al. 2008; Wang and Reinke 2008; Gu et al. 2009; Conine et al. 2010). Ascaris germline maturation, gametogenesis, sexual reproduction, and zygote maturation occur at 37°C in the vertebrate host. We speculate that the adaptation to these processes at 37°C may have altered the role of these temperature-sensitive pathways including the loss of the 21U-RNAs (piRNAs) and 22G-RNAs amplification in the testis. Furthermore, the increased expression and additional targets of Ascaris 26G-RNAs may be an adaptation to the loss or changes in function of 21U- and 22G-RNA pathways. We also failed to detect piRNA pathway components in other nematode parasites of vertebrates, B. malayi and Trichinella, in which spermatogenesis also takes place at 37°C (Ghedin et al. 2007; Mitreva et al. 2011), further suggesting that this may be an adaption to a parasitic life cycle at higher temperatures.
The small RNAs involved in Ascaris spermatogenesis appear to illustrate functional diversification compared with those in C. elegans. Several lines of evidence suggest that C. elegans testis (and embryo soma) 26G-RNAs function as primary siRNAs targeting mRNAs for the synthesis of much more abundant 22G secondary siRNAs (Gent et al. 2009, 2010; Han et al. 2009; Conine et al. 2010; Vasale et al. 2010). During C. elegans spermatogenesis, 26G-RNAs are first produced in distal regions of the testis and subsequently target mRNAs for production of amplified levels of secondary siRNAs in the proximal testis (Fig. 7F). Our analysis of small RNAs throughout the Ascaris male germline (see Fig. 1; Supplemental Table S2) identified distinct expression patterns for 26G- and 22G-RNAs involved in spermatogenesis (Fig. 7F). Although these two small RNA types are both present in the testis, most of Ascaris 26G- and 22G-RNAs have distinct sets of targets: The majority of Ascaris testis 22G-RNAs target nonspermatogenesis-related mRNAs, while Ascaris 26G-RNAs almost exclusively target mRNAs involved in spermatogenesis. For these spermatogenesis-related transcripts, the level of the 26G-RNAs stays high and their ratio to 22G-RNAs associated with these mRNAs is constant throughout spermatogenesis. This reveals a different relationship between 26G- and 22G-RNAs in Ascaris compared with C. elegans for spermatogenesis related genes. Ascaris 26G-RNAs are the major type of testis endo-siRNA targeting mRNAs involved in spermatogenesis. In addition, 26G-RNAs show a bias for the ORF of the mRNA, whereas the less abundant 22G-RNAs show a bias for targeting the 5′ ends of mRNAs (Fig. 7C,F).
Biogenesis of Ascaris endo-siRNAs
Small RNAs produced by RdRPs typically have a 5′ terminal G with polyphosphates (Ambros et al. 2003b; Chicas et al. 2004; Ruby et al. 2006; Pak and Fire 2007; Sijen et al. 2007). 26G-RNAs in C. elegans and Ascaris have a 5′ monophosphate. It has been suggested that C. elegans 26G-RNAs may be derived from dsRNAs formed by RdRPs followed by Dicer cleavage (Han et al. 2009; Conine et al. 2010; Gent et al. 2010; Vasale et al. 2010; Ketting 2011). An alternative hypothesis is that these 26G-RNAs are synthesized by RdRPs with 5′ polyphosphates, followed by trimming of their 5′ terminal phosphates (Ruby et al. 2006). Many questions remain regarding the biogenesis of nematode 26G-RNAs. The absence of the 3′ modification in the Ascaris 26G-RNAs raises a number of questions regarding their biogenesis in Ascaris, and the potential roles of these 3′ modifications in the C. elegans orthologs.
We also observed Ascaris 22G-RNAs in our 5′ monophosphate libraries (Fig. 2G), suggesting Ascaris may also have 5′ monophosphate 22G-RNAs. For each unique Ascaris 22G-RNAs, we found the ratios between 5′ monophosphate and 5′ all-phosphate libraries are constant, suggesting that these monophosphate 22G-RNAs are likely derived from polyphosphate 22G-RNAs (Fig. 6E). Whether these represent intermediates in the turnover or biogenesis of the 22G-RNAs or whether discrete phosphate removal is associated with alternate functions of these small RNAs remains to be determined. We noticed that As.22H RNAs have very similar features to As.22G (Fig. 6) except that they start with different bases, suggesting that As.22H are likely to be generated by RdRPs. Based on their frequency, we speculate that Ascaris RdRPs have a strong preference for G (74%) as the initial base, followed by A (17%), C (7%), and U (2%).
Small RNAs and chromatin diminution
Chromatin diminution eliminates specific DNA sequences from the genome of somatic cells in various organisms, typically during early development (Muller and Tobler 2000; Goday and Esteban 2001). In Ascaris, chromatin diminution occurs during the third through fifth cleavage (four- to 16-cell stage) with the loss of ∼25% of the genome (Muller and Tobler 2000). Fifty percent of the eliminated DNA (∼40 Mb, >300,000 copies) is the highly repetitive satellite DNA that consists primarily of several variants of a 121-bp element (Muller et al. 1982; Streeck et al. 1982).
Elimination of DNA during macronuclear rearrangement in Tetrahymena and other ciliates is associated with small RNAs (Mochizuki and Gorovsky 2004; Yao and Chao 2005). Our initial interest in Ascaris small RNAs was to determine whether small RNAs were mechanistically involved in chromatin diminution. We therefore searched for small RNAs corresponding to the eliminated repetitive DNA in the germline and during early development. We detected small RNAs corresponding to the eliminated satellite DNA in our sequence reads at frequencies of only ∼200/million reads. One would predict that targeting >300,000 copies of a dispersed, repetitive sequence for elimination would require much higher levels of small RNAs, particularly when we observed small RNA reads as high as 1000–250,000 reads/million for endo-siRNAs and miRNAs, respectively. We also did not observe an increase in the expression profiles of small RNAs that target the 121-bp repeats or any small RNAs that were stage or tissue specific that correlated with chromatin diminution (data not shown). We conclude that there is no apparent correlation between small RNAs, the 121-bp repeat, and DNA elimination in Ascaris.
C. elegans CSR-1 Argonaute and 22G-RNAs are required for holocentric chromosome segregation (Claycomb et al. 2009). Claycomb et al. (2009) suggested during chromatin diminution in Ascaris the eliminated regions (e.g., heterochromatic domains and eliminated repeat elements) might not be targeted by small RNAs for retention and chromosome segregation. Chromosome breaks during diminution in the somatic lineages would lead to chromosome fragmentation. Chromosome fragments to be retained would be targeted by CSR-1 Argonaute and 22G-RNAs forming a functional holocentric chromosome, whereas regions for elimination would not be efficiently targeted and not segregated. Experiments are in progress to define the somatic and germline genome in Ascaris to identify the retained and eliminated chromosomal regions and to examine the association of Ascaris CSR-1 Argonaute with these regions.
Methods
Library preparation, sequencing, and reads processing
Small RNA libraries were prepared and sequenced as previously described (Brennecke et al. 2007; Grimson et al. 2008). Computational analysis and annotation of miRNAs, endo-siRNAs, and other cellular RNAs were carried out as described (see Supplemental Material). For genomic libraries, genomic DNA (>50 kb in length) was isolated on CsCl gradients from 32- to 64-cell embryos or from adult tissues using standard methods. Somatic cells predominate in the 32- to 64-cell embryos with only ∼5% germline cells. The A. suum genome was sequenced using a whole-genome shotgun strategy using three sequencing platforms, capillary paired-end Sanger sequence of size fractionated 6-kb cloned sequences and direct 454 titanium and Illumina sequencing (see Supplemental Material).
cDNA libraries were prepared from polyadenylated RNA (2× oligo-dT purified using μMACS mRNA Isolation Kit, Miltenyi Biotec). RNA was treated for 75–150 sec with RNA fragmentation buffer (Ambion) at 70°C to generate RNA fragments 150–400 nt in length. cDNA was prepared by random priming (3–6 μg of random hexamers/1 μg of A+ RNA), and second-stranded cDNA synthesis was carried out using a SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen). Illumina (Solexa) paired-end adaptors were added to the blunt cDNA as described by Illumina, cDNA fragments 300–400 bp in length were gel purified, the fragments were amplified using 17 PCR cycles and paired-end 76- to 100-base sequences were generated as described on the Illumina GAII platform (details of Ascaris samples, library preparation, sequencing, assembly, analysis, and annotation can be found in the Supplemental Materials).
Small RNA labeling and Northern blot analyses
Total RNA was isolated using TRIzol (Invitrogen), and small RNA samples were 5′ labeled by first treating with calf alkaline phosphatase (Roche) followed by phosphorylation with T4 polynucleotide kinase (NEB) and 32P-γ-ATP. Small RNA samples were 3′ labeled using T4 RNA ligase (NEB) and 32P-pCp as described by the manufacturer. RNAs were capped using cold GTP or 32P-α-GTP and guanylyltransferase as described (Cohen et al. 2004).
Northern blots were prepared using 7.5 or 20 μg of total RNA and 5 μg of low-molecular-weight enriched RNA (mirVana miRNA isolation kit with modification). To characterize the 5′ ends of small RNAs, 10 μg of low-molecular-weight enriched small RNAs was treated with Terminator (Epicentre), guanylyltransferase and GTP, or T4 RNA ligase (NEB) and an RNA oligonucleotide. Periodate treatment and β-elimination of RNA were carried out as described (Vagin et al. 2006; Czech et al. 2009). RNAs were separated on denaturing 12.5% or 15% polyacrylamide gels and transferred to Hybond N+ using semi-dry electroblotting (Trans-Blot Semi-Dry transfer cell, BioRad); the membranes were ultraviolet cross-linked and baked at 80°C. Membranes were probed with 5′ end-labeled DNA oligonucleotides.
Bioinformatics
Bioinformatic analyses are described in the Supplemental Material.
Data access
The transcriptome and small RNA data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession numbers GSE26956 and GSE26957. The transcriptome assembly has been submitted to NCBI Transcriptome Shotgun Assembly Sequence Database (http://www.ncbi.nlm.nih.gov/genbank/TSA.html) under accession numbers JI163767–JI182837 and JI210738–JI257410. The genomic sequence reads and assembly data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRP005397 and GenBank accession number AEUI00000000.]
Acknowledgments
We thank Richard Komuniecki, Bruce Bamber, Amanda Korchnak, Vera Hapiak, Jeff Myers, and Routh Packing Co. for their support and hospitality in collecting Ascaris material. We thank David Bartel for helpful discussions and suggestions, Chris Hittinger and Jim Dover for advice on library preparation and sequencing, Tom Evans for samples of C. elegans, and Yingfeng Luo and Songnian Hu at the Beijing Institute of Genomics for computer resources. We also thank Mark Johnston for his comments on the manuscript and the reviewers for their constructive comments and suggestions. B.C. is supported by a PhD fellowship from the Boehringer Ingelheim Fonds. This work was supported in part by grants to R.E.D. (NIH AI0149558 and AI078087), to M.M. (NIH AI081803), and to G.J.H. (grants from the NIH and a kind gift from K.W. Davis). G.J.H. is an investigator of the HHMI.
Footnotes
-
↵4 Corresponding author.
E-mail richard.davis{at}ucdenver.edu.
-
[Supplemental material is available for this article.]
-
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.121426.111.
- Received January 25, 2011.
- Accepted June 8, 2011.
- Copyright © 2011 by Cold Spring Harbor Laboratory Press