Time-series prediction is a common problem in multiple domains for various applications, including retail, industry, smart cities, and financial services. Research in the time-series field is growing exponentially, with hundreds of deep learning time-series forecasting paper submissions to ICML, ECML, ITISE, and multiple journals every year. However, there is currently no common framework to compare the accuracy and performance of all the models from the industry or academia.
- Solution overview
- Setup
- Quick Start Guide
- Advanced
- Memory mapping large datasets
- Running multi-GPU experiments
- Parallel training
- Running experiments with Exponential Moving Averaging
- Running experiments with Curriculum Learning
- Hyperparameter Search
- Custom launchers
- XGBoost Training
- Postprocessing of predictions
- Interprete your model
- Ensembling
- Conversion, Deployment, and Inference
- Parameters
- Release notes
- Reference
Time-Series Prediction Platform (TSPP) enables users to mix and match datasets and models. In this case, the user has complete control over the following settings and can compare side-by-side results obtained from various solutions. These include:
- Evaluation metrics
- Evaluation datasets
- Prediction horizons
- Prediction sliding window sizes
- Model choice
- Model hyperparameters
The platform has the following architecture.
 In the previous figure, the command line feeds the input to the TSPP launcher, which uses said input to configure the components required to train and test the model.
In the previous figure, the command line feeds the input to the TSPP launcher, which uses said input to configure the components required to train and test the model.
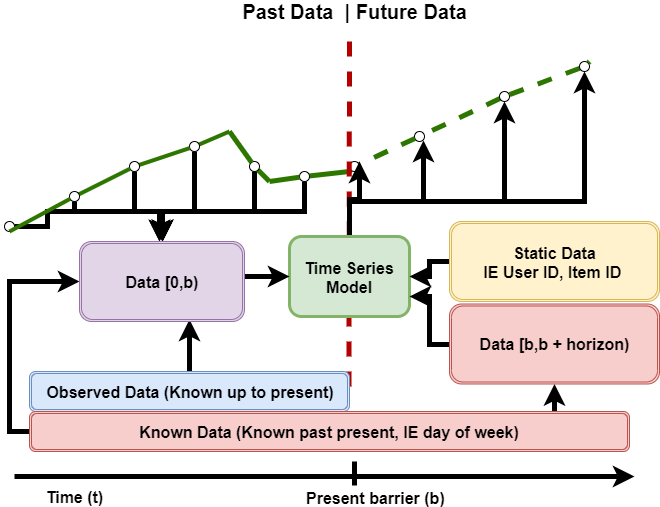
The platform is designed to support multiple data types for input features, including the observed values of the forecasted time-series, known data supporting the forecasts (for example, day of the week), and static data (for example, user ID). This is summarized in the following figure.
This tool supports the following features:
| Feature | Time-Series Prediction Platform |
|---|---|
| Automatic mixed precision (AMP) | Yes |
| Multi-GPU training with (PyTorch DDP) | Yes |
| TorchScript, ONNX, and TRT conversion and NVIDIA Triton Deployment | Yes |
Automatic mixed precision is a mode of computation for PyTorch models that allows operations to use float16 operations instead of float32 operations, potentially accelerating selected operations and total model runtime. More information can be found under the Mixed precision training section.
Multi-GPU training with PyTorch Distributed Data Parallel is a mode of computation for PyTorch models that allows operations to be executed across multiple GPUs in parallel to accelerate computation.
TorchScript, ONNX, and TRT conversion and NVIDIA Triton Deployment refer to the conversion of a model to the aforementioned formats and the ability to deploy the resulting converted models to an NVIDIA Triton inference server. More detail about this process and native inference can be found in the Advanced tab under the Conversion, Deployment, and Inference subsection.
Mixed precision is the combined use of different numerical precisions in a computational method. Mixed precision training offers significant computational speedup by performing operations in half-precision format while storing minimal information in single-precision to retain as much information as possible in critical parts of the network. Since the introduction of Tensor Cores in NVIDIA Volta, and following with both the NVIDIA Turing and NVIDIA Ampere Architectures, significant training speedups are experienced by switching to mixed precision -- up to 3x overall speedup on the most arithmetically intense model architectures. Using mixed precision training requires two steps:
- Porting the model to use the FP16 data type where appropriate.
- Adding loss scaling to preserve small gradient values.
The ability to train deep learning networks with lower precision was introduced in the NVIDIA Pascal architecture and first supported in CUDA 8 in the NVIDIA Deep Learning SDK.
For information about:
- How to train using mixed precision, refer to the Mixed Precision Training paper and Training With Mixed Precision documentation.
- Techniques used for mixed precision training, refer to the Mixed-Precision Training of Deep Neural Networks blog.
- How to access and use AMP for PyTorch, refer to Torch-AMP guide.
Mixed precision can be enabled by specifying trainer.config.amp=True in the launch call. For some cases, when the batch size is small, the overhead of scheduling kernels for mixed precision can be larger than the performance gain from using lower precision, effectively succeeding with lower throughput.
The TSPP utilizes the default configurations provided by each model for each accompanying dataset. More information on individual model configurations can be found within the respective model repositories. By default, Temporal Fusion Transformer (TFT) is included within the TSPP.
- Temporal Fusion Transformers ( conf )
- XGBoost (conf)
- AutoARIMA (conf)
- LSTM (conf)
- N-BEATS (conf)
- N-HITS (conf)
- DeepAR (conf)
- MTGNN (conf)
- DCRNN (conf)
Note: We don't provide scripts to download M5 and PEMS-BAY datasets, each user is responsible for downloading them from corresponding websites.
Note: Each user is responsible for checking the content of datasets and the applicable licenses and determining if suitable for the intended use.
The following section lists the requirements you need to meet to run the Time-Series Prediction Platform.
This repository contains a Dockerfile that extends the PyTorch NGC container and encapsulates some dependencies. Aside from these dependencies, ensure you have the following components:
- NVIDIA Ampere Architecture, NVIDIA Volta or NVIDIA Turing based GPU
- Ubuntu 20.04
- NVIDIA Docker
- Custom Docker containers built for this tool. Refer to the steps in the Quick Start Guide.
For more information about how to get started with NGC containers, refer to the following sections from the NVIDIA GPU Cloud Documentation and the Deep Learning Documentation:
- Getting Started Using NVIDIA GPU Cloud
- Accessing And Pulling From The NGC Container Registry For those unable to set up the required environment or create your own container, refer to the versioned NVIDIA Container Support Matrix.
-
Create a dataset directory. The directory can be arbitrary, and it is recommended not to include it in the TimeSeriesPredictionPlatform directory. This arbitrary directory will be mounted to the TSPP container later. In the following steps, this directory will be referred to as
/your/datasets/. -
Enter the Deep Learning Examples TSPP repository:
cd DeeplearningExamples/Tools/TimeSeriesPredictionPlatform
- Copy the relevant temporal fusion transformer code to the TSPP:
mkdir -p models/tft_pyt/ && cp ../../PyTorch/Forecasting/TFT/modeling.py models/tft_pyt/
- Build the docker image:
docker build -t tspp .
- Next, we will start our container and mount the dataset directory, which means that
/workspace/datasets/points to/your/datasets/. Any changes made to this folder in the docker container are reflected in the original directory and vice versa. If we want to mount additional folders, we can add-v /path/on/local/:/path/in/container/to the run command. This will be useful if we want to save the outputs from training or inference once we close the container. To start the docker container:
docker run -it --gpus all --ipc=host --network=host -v /your/datasets/:/workspace/datasets/ tspp bash
- After running the previous command, you will be placed inside the docker container in the /workspace directory. Inside the container preprocess
electricity,traffic,M5orpems_baydataset. Some of the datasets might need manual download of the data, see datasets:
python data/script_preprocess_data.py --dataset {dataset_name} --output_dir /workspace/datasets/
The raw electricity dataset is the 15-minute electricity consumption of 370 customers from the UCI Electricity Load Diagrams. We aggregate to an hourly forecast and use the previous week to predict the following day. The raw traffic dataset is the 10-minute occupancy rate of San Francisco freeways from 440 sensors downloaded from the UCI PEMS-SF Data Set. We again aggregate to an hourly forecast and use the previous week to predict the following day.
- Preprocess the dataset:
python launch_preproc.py dataset={dataset_name}
- Launch the training, validation, and testing process using the temporal fusion transformer model:
python launch_training.py model=tft dataset={dataset_name} trainer/criterion=quantile
Outputs are stored in /workspace/outputs/{date}/{time}
The TSPP has been designed to work with most CSV sources. To add an arbitrary dataset to the TSPP:
- Enter the Deep Learning Examples TSPP repository:
cd DeeplearningExamples/Tools/TimeSeriesPredictionPlatform
-
Do a preliminary data transposition. TSPP
launch_preproc.pyscript is designed to work with CSV input. Each row should contain only a single datapoint. CSV should contain at least three columns: one for time feature, one for labels, and one for dataset ID (we assume a single file will contain data for multiple correlated time series). For reference, seedata/script_download_data.pyscript. -
Include the target dataset in the directory where you want to keep your datasets. The directory can be arbitrary, and it is recommended not to include it in the TimeSeriesPredictionPlatform directory. This arbitrary directory will be mounted to the TSPP container later
-
Create a configuration file for your dataset, found in TimeSeriesPredictionPlatform/conf/dataset, that includes the following values:
-
source_path: The path to the CSV that contains your dataset
-
dest_path: The path to where preprocessing should write your preprocessed dataset
-
time_ids: The name of the column within your source CSV that is the feature to split your training, validation, and test datasets on.
-
train_range, valid_range, test_range: The ranges that mark the edges of the train, validation, and test subsets. Remember that subsets can overlap, since predicting the first ‘unseen element’ requires the input of the seen elements before it. As an alternative, a
valid_boundarycan be specified, which marks the end of training. Then from thevalid boundary, the nexthorizon lengthnumber of entries are for validation, and finally, from the end of the validation set, the next horizon length number of entries are for testing. -
stride: The stride the dataloader uses to walk the sliding window through the dataset. Default: 1
-
scale_per_id: Whether to scale continuous features during preprocessing using scalers fitted on just samples from the same ID (True), or all samples (False, Default)
-
encoder_length: The length of data known up until the ‘present’
-
example_length: The length of all data, including data known into the future. The prediction horizon is the difference between example_length and encoder_length.
-
features: A list of the features that the model takes as input. Each feature should be represented by an object containing descriptive attributes. All features should have at least a feature_type (ID, TIME, TARGET, WEIGHT, SAMPLE_WEIGHT, KNOWN, OBSERVED, or STATIC) and feature_embed_type (CONTINUOUS or CATEGORICAL). Continuous features may have a scaler attribute that represents the type of scaler used in preprocessing. Categorical columns should have a cardinality attribute that represents the number of unique values the feature takes plus one (this is due to mapping NaNs to 0 in all cases). Examples can be found in the files in /TimeSeriesPredictionPlatform/conf/dataset/. Required features are one TIME feature, at least one ID feature, one TARGET feature, and at least one KNOWN, OBSERVED, or STATIC feature.
-
train_samples: The number of samples that should be taken at train time to use as train input to your model for a single epoch
-
valid_samples: The number of samples that should be taken at train time to use as validation input to your model for a single epoch
-
binarized: Whether preprocessing should accelerate data loading by outputting the preprocessed dataset in a binarized format
-
time_series_count: The number of unique time-series contained in the dataset.
-
-
After a specification has been written, it is ready to be preprocessed with:
docker build -t tspp .
docker run -it --gpus all -v /your/datasets/:/workspace/datasets/ --ipc=host tspp bash
python launch_preproc.py dataset={dataset_name}
For some models, additional parameters are required per dataset. As mentioned in the Adding a new model section, there are examples of these model-dataset combination files in TimeSeriesPredictionPlatform/conf/model_dataset/. An example would be model A requiring a specific hidden size when used on dataset B. In this case, TimeSeriesPredictionPlatform/conf/model_dataset/A_B.yaml should contain the desired hidden size under model.config.hidden_size
- Test your dataset by training and evaluating a Temporal Fusion Transformer. Training, validation, and testing are all included by default using the launch_training.py command shown below:
docker run -it --gpus all -v /your/datasets/:/workspace/datasets/ --ipc=host tspp bash
python launch_training.py dataset={YOUR_DATASET} model=tft trainer/criterion=quantile
see example
In this section, we will demonstrate how to add a new dataset. Let's assume we want to add a few sine waves dataset to demonstrate the model's ability to fit a deterministic timeseries, in TSPP we should follow these steps:- Create dataset and save in datasets directory
# script_generate_sine.py
import os
import numpy as np
import pandas as pd
if __name__ == '__main__':
dest_path = '/workspace/datasets/sines/'
os.makedirs(dest_path, exist_ok=True)
# generate series with general form y = k * sin(x)
ks = [1, 10, 125]
xs = np.linspace(0, 4*np.pi, num=200)
df = pd.concat([pd.DataFrame({'y': k * np.sin(xs), 'x': xs, 'point_idx': np.arange(len(xs)), 'ts_id': i}) for i, k in enumerate(ks)])
df.to_csv(os.path.join(dest_path, 'sines.csv'))python script_generate_sine.py- Create dataset description for data in
conf/dataset/<name>.yamlFor our example we want to predict the next value based on series id, previous value and corresponding x values.example_lengthis going to be 2, since we want to predict the next value from previous andencoder_lengthis 1 to indicate that 1 of 2 values in the example are used as a lookback window.
For evaluation and testing, we will leave last two values. Easiest way to achieve this is to set valid_boundary (to learn more on how to use ranges instead of boundary, refer to electricity.yaml).
Also, since we know that series have widely different scales, we want to scale them separately, hence scale_per_id
# conf/dataset/sines.yaml
_target_: data.datasets.create_datasets
config:
graph: False
source_path: /workspace/datasets/sines/sines.csv
dest_path: /workspace/datasets/sines/
valid_boundary: 198
scale_per_id: True
encoder_length: 1
input_length: 1
example_length: 2
stride: 1
MultiID: False
features:
- name: 'ts_id'
feature_type: 'ID'
feature_embed_type: 'CATEGORICAL'
cardinality: 4
- name: 'point_idx'
feature_type: 'TIME'
feature_embed_type: 'CONTINUOUS'
- name: 'y'
feature_type: 'TARGET'
feature_embed_type: 'CONTINUOUS'
scaler:
_target_: sklearn.preprocessing.StandardScaler
- name: 'x'
feature_type: 'KNOWN'
feature_embed_type: 'CONTINUOUS'
scaler:
_target_: sklearn.preprocessing.StandardScaler
- name: 'ts_id'
feature_type: 'STATIC'
feature_embed_type: 'CATEGORICAL'
cardinality: 4
train_samples: -1
valid_samples: -1
binarized: True
time_series_count: 3NOTE: How the same column can be used to describe several different features. The only one you probably wouldn't want to appear more than once is TARGET column ;)
- Congratulations! You created your own dataset, now you can proceed to preprocess it and train your models with it.
python launch_preproc.py dataset=sines
python launch_training.py dataset=sines model=tft trainer.config.num_epochs=1Models added to the prediction platform are subject to a few key constraints. Namely, the models should be constructed using PyTorch. Models should handle the forecasting task (anomaly detection and classification are planned); models should expect that the data is fed in a sliding window and that tensors will be aggregated by Temporal/Data Type. An example of this can be found in data/dataset.py.
The default format of the data batch is a dictionary with tensors representing different kinds of covariates. A complete list of the tensor names and types that can be found in a batch:
FEAT_NAME_MAP = {
"s_cat": (InputTypes.STATIC, DataTypes.CATEGORICAL),
"s_cont": (InputTypes.STATIC, DataTypes.CONTINUOUS),
"k_cat": (InputTypes.KNOWN, DataTypes.CATEGORICAL),
"k_cont": (InputTypes.KNOWN, DataTypes.CONTINUOUS),
"o_cat": (InputTypes.OBSERVED, DataTypes.CATEGORICAL),
"o_cont": (InputTypes.OBSERVED, DataTypes.CONTINUOUS),
"target": (InputTypes.TARGET, DataTypes.CONTINUOUS),
"weight": (InputTypes.WEIGHT, DataTypes.CONTINUOUS),
"sample_weight": (InputTypes.SAMPLE_WEIGHT, DataTypes.CONTINUOUS),
"id": (InputTypes.ID, DataTypes.CATEGORICAL),
"timestamp": (InputTypes.TIME, DataTypes.CATEGORICAL) # During preprocessing we cast all time data to int
}
To integrate a model into the TSPP:
- Enter the Deep Learning Examples repository:
cd DeeplearningExamples
- Copy the model files into the Deep Learning Examples DeepLearningExamples/Tools/PyTorch/TimeSeriesPredictionPlatform/models/ directory:
cp -r /PATH/TO/YOUR/MODEL Tools/PyTorch/TimeSeriesPredictionPlatform/models
- Write a configuration file for the model in
DeepLearningExamples/Tools/TimeSeriesPredictionPlatform/conf/model.
This configuration file should reflect the default configuration for your model. Within this file, the target of the model component should be set to point to your model class and global override to define the trainer type used for your model. Currently, we support ctltrainer - used for DL models, stat and xgb trainers used for stat and xgb models respectively. If you need a custom trainer, you can check conf/trainer/<type>trainer.yaml or feel free to open an issue with suggestions. For example, on how to define a trainer for your model, see conf/model/tft.yaml.
If your model needs additional configuration values based on the dataset, you should create a configuration file in DeepLearningExamples/Tools/TimeSeriesPredictionPlatform/conf/model_dataset/{modelname_datasetname.yaml} named according to the model and dataset names. Examples can be found in the DeepLearningExamples/Tools/TimeSeriesPredictionPlatform/conf/model/tft.yaml and DeepLearningExamples/Tools/TimeSeriesPredictionPlatform/conf/model_dataset/tft_traffic.yaml files.
- Build and launch container:
cd DeepLearningExamples/Tools/TimeSeriesPredictionPlatform
docker build -t tspp .
docker run -it --rm --ipc=host --network=host --gpus all -v /your/datasets/:/workspace/datasets/ tspp bash
- Verify that the model can be run within the TSPP:
python launch_training.py model={model_name}
Some additional values may be needed in this call. For example, if your model requires the Gaussian NLL criterion, you will need to append trainer/criterion=GLL to your call.
see example
Let's assume that you want to test linear model performance for your research and you want to consume all static categorical and known continuous data you have available: 1. Write your model that consumes `config` argument:# models/linear.py
import torch
import torch.nn as nn
class LinearModel(nn.Module):
def __init__(self, config):
super().__init__()
self.encoder_length = config.encoder_length
self.example_length = config.example_length
self.forecest_len = self.example_length - self.encoder_length
self.num_targets = config.temporal_target_size
self.input_size = (len(config.static_categorical_inp_lens) +
config.temporal_known_continuous_inp_size) * self.encoder_length
self.model = nn.Linear(self.input_size,
self.num_targets * self.forecest_len,
bias=config.use_bias)
def forward(self, batch):
batch_size = batch['target'].shape[0]
inp = torch.cat((batch['s_cat'][:, :self.encoder_length].view(batch_size, -1),
batch['k_cont'][:, :self.encoder_length].view(batch_size, -1)),
dim=1)
pred = self.model(inp)
return pred.reshape(batch_size, self.forecest_len, self.num_targets)- Write
conf/model/entry for your model:
# conf/model/linear.yaml
_target_: models.linear.LinearModel
config:
use_bias: True
defaults:
- _self_
- /trainer@_global_/trainer: ctltrainerNOTE: static_continuous_inp_size, temporal_known_continuous_inp_size, etc. are 'magic' values that are injected into config during parsing time, to know more see conf/train_derived_fields.yaml. This file connects different parts of config and creates aggregate fields that, for example, keep track of number and cardinality of categorical variables in model config.
- Congratulations! You are ready to train your model.
python launch_training.py model=linear dataset=electricityThe following sections provide more details about changing the dataset, altering the data preprocessing, and comparing the training results.
Since March 2024 release, we have an option designed for large datasets. Instead of loading dataset into RAM, you can use option +dataset.config.memory_mapped=True with launch_training.py to memory map dataset from the drive. Note, however, that in order to saturate GPUs, you will have to increase the number of dataloader workers trainer.config.num_workers to compensate for longer example loading time. Loading time depends heavily on drives and file system your machine uses.
Launching on multi-GPU requires no changes to model code and can be executed as follows within a TSPP container:
python launch_training.py -m hydra/launcher=torchrun hydra.launcher.nproc_per_node={num_gpus} {override parameters}
Statistical models (like AutoARIMA) do not run on GPU, so they are unsuitable for multi-GPU acceleration. In addition, XGBoost has a separate way of doing multi-GPU acceleration (see dask_xgboost).
While doing seed sweeps or hp searches on a machine with more than one GPU, we can parallelize the workload by using the joblib hydra plugin. To use the plugin, one has to specify hydra/launcher=joblib together with the number of parallel jobs hydra.launcher.n_jobs=8. For example:
python launch_training.py \
-m \
seed='range(1,17)' \
model=tft \
dataset=electricity \
trainer/criterion=quantile \
trainer.config.num_epochs=3 \
hydra/launcher=joblib \
hydra.launcher.n_jobs=8 \
hydra.sweeper.max_batch_size=8Warning: Sweeper sends jobs to a launcher in batches. In order to avoid race conditions, specify sweeper batch size to exactly match the number of parallel jobs. For the default sweeper it would be: hydra.sweeper.max_batch_size=8, and for optuna sweeper: hydra.sweeper.n_jobs=8.
Exponential moving averaging is a technique in which, while training, the model weights are integrated into a weighted moving average, and the weighted moving average is used in lieu of the directly trained model weights at test time. Our experiments have found this technique improves the convergence properties of most models and datasets we work with. The full paper of EMA can be found here.
To activate EMA in the TSPP, specify trainer.config.ema=True in the command line call at runtime. The decay parameter in the moving average can be modified using the +trainer.config.ema_decay={decay}.
To use curriculum learning in your training, specify trainer.config.cl_start_horizon and trainer.config.cl_update config fields. More on CL
Hyperparameter searches can be used to find close-to-optimal hyperparameter configurations for a given model or dataset. In the TSPP, hyperparameter searches are driven by Optuna. To launch a hyperparameter search, use:
python launch_training.py -m hydra/sweeper=optuna hydra.sweeper.n_trials={N} {parameter_ranges}For example: let's tune model size and learning rate for tft model on electricity dataset.
export RESULTS=/ws/test_sweep
mkdir -p ${RESULTS}
python launch_training.py -m \
'model.config.n_head=choice(1,2,4)' \
'model.config.hidden_size=choice(128,256)' \
'trainer.optimizer.lr=tag(log, interval(1e-5, 1e-2))' \
model=tft \
dataset=electricity \
trainer.config.batch_size=1024 \
evaluator.config.batch_size=1024 \
hydra/sweeper=optuna \
+optuna_objectives=[MAE,RMSE] \
hydra.sweeper.direction=[minimize,minimize] \
hydra.sweeper.n_trials=128 \
hydra/launcher=joblib \
hydra.launcher.n_jobs=8 \
hydra.sweeper.storage="sqlite:///${RESULTS}/hp_search_multiobjective.db"For more info how to properly set up {parameter_ranges} visit hydra docs
TSPP now have custom sweeper and launchers in order to boost performance during hp searches with optuna. To utilize more of your resources during long sweeps, you can select multiprocessing as your launcher and set your hydra.sweeper.experiment_sequence to hydra_utils.TSPPOptunaExperimentSequence in the existing sweeps:
python launch_training.py -m \
hydra/sweeper=optuna \
hydra/launcher=multiprocessing \
hydra.launcher.n_jobs=<num_gpus> \
hydra.sweeper.n_trials={N} \
hydra.sweeper.experiment_sequence=hydra_utils.TSPPOptunaExperimentSequence \
{parameter_ranges}This might boost performance, especially if you are using early stopping and sweep over model sizes or any other parameter that changes model training time. For more information and motivation behind the changes, see this hydra issue and related PR
XGBoost and RAPIDS packages are now automatically present in the base NGC PyTorch containers. The TSPP is able to leverage this and allow users to perform training, inference, and deployment on XGBoost and Dask XGBoost using the same commands as neural network models. To train:
python launch_training.py model={xgboost, dask_xgboost} dataset={dataset}Note: All stages of XGBoost are run on GPU. CPU training is currently not supported.
This launches training using CSV files from the output of preprocessing. Validation data is automatically used for early stopping if applicable.
The TSPP trains a separate XGBoost model for each step in the horizon. If some arbitrary row in the dataframe is at time t, then for the ith model, we train it to predict timestep t+i. As a part of this, we give the model access to all the features at time step t and bring up the static and known features at timestep t+i. Each ID is handled separately, so for any given training/prediction sample, there is only data from 1 ID.
XGBoost itself cannot create new features or process features in the same way as neural networks. To this end, we have created a framework where one can specify lag_features and moving_average_features. lag_features allow the XGBoost model to have access to the values of the given feature in the past, while moving_average_features allow the model to have access to the moving average of the given feature to some number of previous time steps. For an example of how to specify these features, take a look at conf/model_dataset/xgboost_electricity.yaml. To specify a lag_feature, one needs to select a feature, a min value, and a max value. The TSPP then automatically adds the values of that feature at timestep t-min_value to t-max_value. Instead of specifying min and max, one can also specify value, which is a list of values for finer control. Note the values must be greater than 0 and must be natural numbers.
To specify a moveing_average_feature, one needs to select a feature and a window_size. This window_size indicates that a new feature will be added that is the average of the values of the feature from t-window_size to t.
For model parameters, the standard XGBoost parameters can be passed using model.config.{parameter}, some may require +model.config.{parameter} if the parameter is not set inside the conf/ directory. In addition, one can specify the number of boosting rounds using model.config.n_rounds.
There are a few additional parameters that are used exclusively for DaskXGBoost for initialization of the LocalCUDACluster: model.config.cluster.world_size, which sets the number of GPUs to use, model.config.cluster.device_pool_frac, which sets the amount of memory to allocate on the GPUs, model.config.cluster.protocol which sets the protocol to use on the cluster, and model.config.cluster.npartitions which sets the number of partitions to use for converting to Dask-cuDF.
Finally, trainer.callbacks.early_stopping.patience can be used to set the early stopping patience of the XGBoost rounds, and trainer.config.log_interval can be used to set the frequency of the logging for XGBoost.
Some datasets require additional post-processing to make predictions more accurate, e.g. during sales prediction you are more than sure to sale at least 0 of a given product. That's why TSPP evaluator module now support postprocessing. To use it, you need to set evaluator/postprocessor to one of the predefined postprocessors, or create your own in conf/evaluator/postprocessor/
python launch_training.py model=tft dataset=electricity trainer.config.num_epochs=1 evaluator/postprocessor=clip_to_zeroFor selected models (ones that inherit from InterpretableModelBase) TSPP allows users to visualize activations/attention masks/etc. In order to visualize the model, you have to add TensorBoardBackend and/or WandBBackend to your conf/logger.yaml, since those backends support figure logging. Next, you need to specify which examples you want to visualize adding +evaluator.config.visualisation_indices to your config. For example to visualise tft attention on samples 1, 1025, 1026 and 2048 use:
TFT_SCRIPTING=1 python launch_training.py \
dataset=${DATASET} \
model=tft \
trainer/criterion=quantile \
trainer.config.batch_size=1024 \
+evaluator.config.visualisation_indices='[1, 1025, 1026, 2048]'Note: Interpretability for TFT model requires environmental variable TFT_SCRIPTING to be set.
Currently, we only support ensembling of DL models with the same type. This can be used to ensemble same models with different hyperparameters, most commonly random generator seed. For example, this script trains 8 models with different seeds and then uses them to produce one prediction:
RESULTS=/ws/tft_ensemble_checkpoints
mkdir -p ${RESULTS}
python launch_training.py \
-m \
seed="range(1,9)" \
model=tft \
dataset=electricity \
overrides=tft/electricity/best_0 \
trainer.config.log_interval=-1 \
+trainer.config.force_rerun=True \
evaluator.config.metrics=[MAE,RMSE,SMAPE,TDI] \
hydra.sweep.dir=${RESULTS} \
hydra/launcher=joblib \
hydra.launcher.n_jobs=8 \
hydra.sweeper.max_batch_size=8
rm ${RESULTS}/*/last_checkpoint.zip
MODEL_LIST="["
for I in $( seq 0 7 )
do
MODEL_LIST+="{dir: ${RESULTS}/$I, checkpoint: best_checkpoint.zip, weight:1.0},"
done
MODEL_LIST=${MODEL_LIST::-1}
MODEL_LIST+="]"
TFT_SCRIPTING=1 python launch_ensembling.py model.config.model_list="${MODEL_LIST}"Note: we export TFT_SCRIPTING to use native torch.nn.LayerNorm instead of apex.FusedLayerNorm. Using apex implementation might lead to errors during ensembling and it will be removed in the next release as this API is deprecated.
Inference takes place after a model has been trained and one wants to run data through. Since this only entails using a forward function, the model can be optimized and converted to many different formats that can perform the forward pass more efficiently. In addition, one can set up a NVIDIA Triton inference server, which allows for a continuous stream of data to be presented to and passed through the model. The server provides an inference service via an HTTP or gRPC endpoint at ports 8000 and 8001, respectively, on the “bridge” docker network.
The TSPP supports a few versions of inference, including native inference and NVIDIA Triton deployment. Both use the test_forward function specified in the model config (defaults to forward()) as the forward function.
To launch native inference, one must have a checkpoint directory from a TSPP training call that includes a .hydra directory and a best_checkpoint.zip from training a Neural Net, a populated checkpoints directory from training an XGBoost, or an arima.pkl file from training an ARIMA model. Then run
python launch_inference.py checkpoint=/path/to/checkpoint/directory
Note: Do not confuse the checkpoint directory with the TimeSeriesPredictionPlatform/outputs/ directory. The directory to use in the inference call is typically two levels lower (for example, /path/to/TimeSeriesPredictionPlatform/outputs/2021-08-23/03-03-11/).
The device argument refers to the device that one would like the model to be built on and run on. Note that multi-GPU inference launches are not supported. By default, the evaluator uses the configs specified in the .hydra/config.yaml file from the checkpoint directory. One can override these by including them in the launch. For example, if one wanted to adjust the metrics to use MAE and RMSE only.
python launch_inference.py checkpoint=/path/to/checkpoint/directory “+inference.config.evaluator.config.metrics=[‘MAE’, ‘RMSE’]”
Note: Be sure to include the + when necessary, this special character will add new fields to the current config. However if value already exists, this will result in error from hydra.
Prior to the next section, make sure that the TSPP container is run with the following arguments from the TSPP directory. We recommend an outputs_dir is created that can be used to mount the outputs directory and the multirun folder from multi-GPU runs.
docker run -it --rm --gpus all --ipc=host --network=host -v /your/datasets/:/workspace/datasets/ -v /your/outputs_dir/:/your/outputs_dir/ -v $(pwd):$(pwd) -v /your/outputs_dir/outputs/:/workspace/outputs/ -v /your/outputs_dir/multirun/:/workspace/multirun/ -v /var/run/docker.sock:/var/run/docker.sock tspp
Note that /your/outputs_dir/{outputs/multirun} is equivalent to the python script os.path.join(/your/outputs_dir/, outputs).
In the previous command, note that six different directories are mounted. The datasets are mounted to the usual location, but we have two different mount locations for outputs. Mounting the outputs to /workspace/outputs/ allows usual training calls to be saved in your output directory. Similarly, mounting the multirun to /workspace/multirun/ allows multi-GPU to be saved. The second output mount is mounted to the same path as the output directory is in the host. This is essential due to the way we deploy to NVIDIA Triton. The directory of the output in the docker must match the directory of the output on the host machine. Additionally, the mount for /var/run/docker.sock allows the tspp docker container to launch another container. In our case, this is the NVIDIA Triton server. In subsequent calls to launch_triton_configure.py, the /path/to/checkpoint/directory/ must be of the form /your/outputs_dir/{checkpoint_dir} instead of /workspace/{checkpoint_dir} and should be absolute paths.
Remember that multi-GPU runs are stored in multirun instead of outputs.
To use deployment, the simplest way is to use the directories multirun and outputs directly inside the TSPP. This can be achieved by launching the docker as follows.
docker run -it --rm --gpus all --ipc=host --network=host -v /your/datasets/:/workspace/datasets/ -v $(pwd)/multirun:/workspace/multirun -v $(pwd)/outputs:/workspace/outputs -v $(pwd):$(pwd) /var/run/docker.sock:/var/run/docker.sock tspp
Finally, note that to run the deployment script, you must be in the same directory path in the container as the TSPP is stored on your machine. This means that being in /workspace in the container may not work for running the deployment. If outside the container your TimeSeriesPredictionPlatform is at /home/user/TimeSeriesPredictionPlatform, you must be at the same path in your docker container (/home/user/TimeSeriesPredictionPlatform). This is the purpose of the -v $(pwd):$(pwd) in the run script.
To launch conversion and deployment, one must again have a checkpoint directory from a TSPP training call that includes a .hydra directory and a best_checkpoint.zip from a Neural Net training or a populated checkpoints directory from an XGBoost training. Stats model, such as Arima, are not supported for deployment. In addition, the model that will be converted must already support conversion to the required format. In the current version of the TSPP, we first export the model to either TorchScript-Script or TorchScript-Trace and subsequently convert it to TorchScript, Onnx, or TRT using the model-navigator package. We also support export to Onnx and conversion to both Onnx and TRT. For XGBoost models, we format the checkpoints and deploy using the FIL backend; there are no extra steps necessary. To run export and conversion (for XGBoost, the deployment/export and deployment/convert fields can be ignored, and no other deployment options are functional):
python launch_triton_configure.py deployment/export={ts-trace, ts-script, onnx} deployment/convert={torchscript, onnx, trt} checkpoint=/path/to/checkpoint/directory
The format mapping is listed below:
- TorchScript-Script: ts-script
- TorchScript-Trace: ts-trace
- TorchScript: torchscript
- Onnx: onnx
- TRT: trt
Note that the conversions do not support the apex fused LayerNorm library. In order to get around this, we set the os environ variable ‘TFT_SCRIPTING” to True when loading the model for deployment. This changes the apex LayerNorm to vanilla torch LayerNorm. In addition, one can select the batch size and precision of the conversion, using +inference.config.evaluator.config.batch_size and inference.config.precision=Choice[ fp32, fp16 ] respectively. Once export and conversion have been done, the results are stored in /path/to/checkpoint/directory/deployment. Subsequently, the converted model’s NVIDIA Triton config is generated in the /path/to/checkpoint/directory/deployment/navigator_workspace/model-store/ directory. An additional option in running conversion is selecting whether to run the basics of conversion and NVIDIA Triton config creation or to run the full pipeline of conversion, NVIDIA Triton config creation, profiling, analysis, and helm chart creation. Setting config.inference.optimize=True during launch switches to the full pipeline. Another part of optimization is setting the backend accelerator for NVIDIA Triton config generation. Setting config.inference.accelerator=Choice[none, trt] changes the accelerator specified. Note that this defaults to ‘none’ and ‘trt’ is only compatible with the Onnx conversion. If one wants to launch the NVIDIA Triton inference server using a specific GPU, the CUDA index can be specified with the config option config.inference.gpu, which defaults to 0.
More information on the conversion is located here
More information on the NVIDIA Triton config creation is located here
More information on the full pipeline is located here
After running launch_triton_configure.py, the directories are set up for quick Triton deployment. To start the server:
python launch_inference_server.py checkpoint=/path/to/checkpoint/directory
Once the script finishes running, the Triton server will run in the background waiting for inputs until it is closed. In order to run inference on the test dataset, the checkpoint was trained on:
python launch_inference.py inference=triton checkpoint=/path/to/checkpoint/directory
Similar to the native inference, one can again override the evaluator configs. The NVIDIA Triton model name is set as the second directory to the model. For example, in the case of our TFT model, whose path is models.tft_pyt.TemporalFusionTransformer, the name of the NVIDIA Triton model is tft_pyt. In the case of XGBoost, there is a different model name for each model in the horizon length, specified as xgb_{i}.
There is a config option +inference.config.model_name, which can be set to the NVIDIA Triton model name. This does not set the name of the model but instead selects which of the possible models in the model-store directory will be used for inference. This is useful after a call using the optimize option, which can generate multiple different models in the model-store.
For both the native and triton launch_inference, one can specify what dataset and target_scalers to use (if any) as long as the data shapes do not conflict with the already trained model. To specify a dataset directory use +inference.config.dataset_dir=/path/to/dataset. The dataset directory must contain a tspp_preprocess.bin file as well as either train.bin/valid.bin/test.bin or train.csv/valid.csv/test.csv, depending on the configuration option dataset.config.binarized (this option cannot be changed during deployment or inference). Once the path has been set, deployment and inference both use the test dataset.
The TSPP also supports an online inference solution for both XGBoost models and Neural models. Given raw data (not preprocessed by TSPP), both native and NVIDIA Triton inference can preprocess and pass the data through the models. When running, specify +inference.config.dataset_path=/path/to/raw/data/csv and if applicable +inference.config.preproc_state_path=/path/to/tspp_preprocess.bin (if the preprocess state is saved elsewhere). Note this is not yet supported on ARIMA models.
As a final note, make sure to close the NVIDIA Triton Inference Server docker container when finished using docker stop trt_server_cont.
Our TFT model supports export to TorchScript-Trace and conversion to all formats.
If you encounter an error such as
RuntimeError: Model tft_pyt:1 is not ready
Or
ERROR root Exception in callback <function InferenceServerClient.async_infer.<locals>.wrapped_callback at 0x7f9437b469d0>: AttributeError("'InferenceServerException' object has no attribute 'get_response'")
There are a few possible reasons for this to come up. First, make sure that when the TSPP docker container was launched, the network argument was set to host. Second, ensure the correct initial path is used, so something of the form /home/user/TimeSeriesPredictionPlatform instead of /workspace. Next, one can run “docker ps”; if the container “trt_server_cont” shows up, close it using “docker stop trt_server_cont”. After this, one should try rerunning the command. If neither of these steps is applicable or the problem persists, it is a more specific issue that requires more debugging.
Config structure reflects the internal design of the tool. Most components have their config stored in
/workspace/conf/{component_type}/{component_name}.yaml
With a few exceptions where components are strictly dependent (for example, optimizer can be used only during training, so its configuration is stored in /workspace/conf/trainer/optimizer/{optimizer_name}.yaml)
If a parameter does not exist in the config, you must prepend + to its reference in the command line call. For example, +trainer.config.force_rerun=... adds force_rerun to trainer.config, but trainer.config.force_rerun=... errors.
March 2024
- Added memory mapped datasets
- Added ensembling module
- Added wandb logging
- Added postprocessor
- Added timestamps to predictions
- Added visialization and interpretability
- Added custom hydra plugins
- Added support for models from the paper
- Added support for dataset from the paper
July 2022
- Reworked config structure
- Added parallel execution
- Fixed race condition when using torch distributed
- Switched to optuna plugin instead of having custom code
- Added basic suspend resume utility
- Added curriculum learning option
- Weights are allowed for arbitrary loss function
- Removed visualization (will be added in a future release)
- Added XGBoost model
- Added multi ID dataset for models like Informer
- Added example scripts
- Criterions and optimizers no longer require dummy wrappers
November 2021
- Initial release
If you encounter errors stating srcIndex < value, verify that your categorical cardinalities are the correct size, this indicates that the value of a categorical you are trying to embed is too large for its respective embedding table.
Cite the following paper if you find this code useful or use it in your own work:
@misc{bączek2024tspp,
title={TSPP: A Unified Benchmarking Tool for Time-series Forecasting},
author={Jan Bączek and Dmytro Zhylko and Gilberto Titericz and Sajad Darabi and Jean-Francois Puget and Izzy Putterman and Dawid Majchrowski and Anmol Gupta and Kyle Kranen and Pawel Morkisz},
year={2024},
eprint={2312.17100},
archivePrefix={arXiv},
primaryClass={cs.LG}
}