Abstract
Gaining clinicians’ trust will unleash the full potential of artificial intelligence (AI) in medicine, and explaining AI decisions is seen as the way to build trustworthy systems. However, explainable artificial intelligence (XAI) methods in medicine often lack a proper evaluation. In this paper, we present our evaluation methodology for XAI methods using forward simulatability. We define the Forward Simulatability Score (FSS) and analyze its limitations in the context of clinical predictors. Then, we applied FSS to our XAI approach defined over an ML-RO, a machine learning clinical predictor based on random optimization over a multiple kernel support vector machine (SVM) algorithm. To Compare FSS values before and after the explanation phase, we test our evaluation methodology for XAI methods on three clinical datasets, namely breast cancer, VTE, and migraine. The ML-RO system is a good model on which to test our XAI evaluation strategy based on the FSS. Indeed, ML-RO outperforms two other base models—a decision tree (DT) and a plain SVM—in the three datasets and gives the possibility of defining different XAI models: TOPK, MIGF, and F4G. The FSS evaluation score suggests that the explanation method F4G for the ML-RO is the most effective in two datasets out of the three tested, and it shows the limits of the learned model for one dataset. Our study aims to introduce a standard practice for evaluating XAI methods in medicine. By establishing a rigorous evaluation framework, we seek to provide healthcare professionals with reliable tools for assessing the performance of XAI methods to enhance the adoption of AI systems in clinical practice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Artificial intelligence (AI) and, in particular, machine learning (ML) may potentially revolutionize healthcare if machine learning-based predictors (MLBPs) will become transparent enough to gain the trust of clinicians and, possibly, of patients [1,2,3]. However, many ML models, such as deep neural networks, lack in transparency and are often referred to as “black boxes”. Their predictions are based on mathematical models exploiting complex patterns in data that pose challenges in terms of comprehension. In healthcare, where decisions can have life-altering consequences, it is imperative to explain to clinicians why MLBPs suggest particular recommendations or diagnoses.
Explaining decisions is seen as the solution to increase trust in MLBPs, and, consequently, explainable artificial intelligence (XAI) has emerged as a prominent area of research in the context of medical applications [2,3,4]. XAI represents a broad trend in the computational community, aiming to shed light on the underlying mechanisms of ML algorithms, thus providing insights into their decision-making processes. XAI approaches encompass diverse techniques and methodologies including feature importance analysis, rule extraction, surrogate models, and visualization techniques. These methods strive to identify the most influential features or patterns in data that significantly contribute to the predictions of such ML models. By observing these influential features or patterns, clinicians can assess the reliability and validity of MLBPs, that enable them to make well-informed decisions based on the provided insights.
However, the mere explanation of the decisions relying on MLBPs is insufficient to build trust in clinicians. In fact, XAI models should be carefully evaluated to determine their utility [5, 6]. Evaluating XAI models is extremely difficult as evaluation methods should be tailored to users and not to system developers. Indeed, the goodness of an explanation model depends on one important factor: the knowledge of users. Moreover, evaluating XAI models should be as objective as possible.
The major contributions of this paper are the following:
-
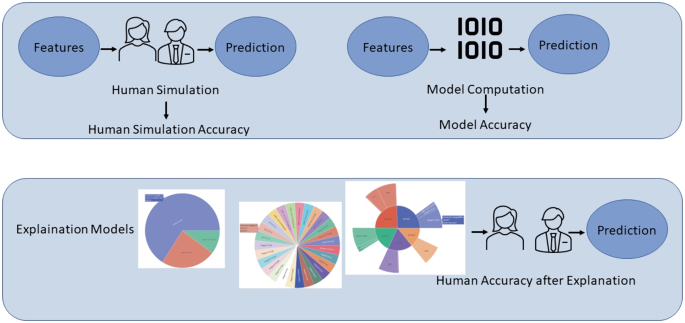
Proposing an evaluation procedure for XAI models for clinicians (see Fig. 1). Our procedure applies forward simulatability [7, 8] to the context of clinical practice by overcoming its limitations. Forward simulatability aims to evaluate the mental model of clinicians built by using explanations [9]. Indeed, it assigns a score to an explanation model by measuring the ability of a human predictor to replicate machine learning decisions when exposed to the provided explanations. To the best of our knowledge, this is the first attempt to apply forward simulatability to the medical domain.
-
Creating three explanation models for the ML-RO system [10]. This system is an MLBP built on support vector machines (SVMs) with random optimization to extract assigned weights to groups of features and the features within each group. The specific nature of ML-RO allows us to define different XAI models based on the importance of the features that can be compared by using forward simulatability.
-
Performing an assessment of the evaluation procedure of XAI models on three challenging clinical datasets.
The rest of the paper is organized as follows. The “Background” section analyzes the current XAI models and the notion of forward simulatability. The “Methods” section presents the evaluation procedure for XAI, analyzing the limitation of forward simulatability in the medical domain. The “Challenging Clinical Datasets” section presents the three challenging systems and the related XAI methods. The “Results and Discussion” section discusses results. Finally, the “Conclusions” section draws the conclusions of our study.
Background
Explaining decisions of machine learning algorithms is a crucial yet difficult objective, and clear evaluation strategies are needed to assess the relative quality of explanation methods. Explainability, often called interpretability [11, 12], is a flourishing field as machines, which learn from data, may make good decisions for obscure reasons. Hence, a strategy to understand which explanation method is better than the others is necessary. This field is generally referred to as XAI.
XAI models are divided into two main categories: model-centric and post hoc [13, 14]. Model-centric XAI approaches aim to explain the learned model itself [15,16,17]. In this sense, decision trees [18] learned from data are one of the most transparent machine learning models. Conversely, post hoc XAI approaches aim to explain a model by providing verbal or visual explanations [15, 19]. Post hoc visual explanations, previously used in the medical domain [2, 20], are graphs, charts, and saliency heatmaps. In the medical field, post hoc explanations are extremely important as these explanations may help clinicians to gain trust in MLBPs during their clinical practice.
An evaluation strategy of XAI methods should capture trust, causality, and informativeness [15]. Trust can be defined as a combination of various aspects: trust in the model performance, trust in the model accuracy, subjective perception by the user, and the similarity between model behavior and human behavior in making decisions (i.e., the model makes mistakes in the same cases of human mistakes). Causality is related to the capability of inferring proprieties of the natural world by machine learning models. In medicine, this is a crucial aspect of artificial intelligence models because it makes it possible to reveal a strong association between features and events. Likewise, an explanation should be informative. Human decision-makers should be provided with information that can be useful for them. Informativeness concerns the capability of machine learning and explanation models to provide valuable information regarding the decision process. In the field of medicine, it is crucial to have an XAI method that increases the trust in MLBPs by explaining the causality and being informative.
However, an evaluation strategy of XAI models poses an additional, important challenge as explanations may be evaluated only if understood [6]. Indeed, readers of explanations should have the correct level of culture. An application-grounded evaluation [21] is seen as the solution. In this approach, consumers of the final application are considered the best candidates to perform the evaluation as they should be experts in the application domain. In our case, clinicians are the final users of the application. Therefore, post hoc explanations should be tailored to them and not to computer scientists.
Forward simulation or simulatability [7] is a clever way for post hoc evaluation of XAI models in an application-grounded evaluation scheme. In forward simulation, experts are asked to make decisions by using the explanations of the decisions of MLBPs. Then, these decisions are compared with the decisions of MLBPs. The assumption is that if experts produce the same outcome of MLBPs, explanations are effective. Forward simulation has been extensively applied to rigorously evaluate explainability [8] in comparing different interpretability algorithms such as LIME [22], and ANCHORS [23] on the text and tabular data. These algorithms are designed to determine the importance of features in decision-making. However, to the best of our knowledge, forward simulation has never been applied in the medical domain. The medical domain has specific characteristics that may hinder the applicability of this particular technique for evaluating explanation systems. In this study, we aim to apply this evaluation scheme to the medical domain, analyze its limits, and propose mitigation measures.
Methods
In this section, we propose our method to evaluate XAI models in the clinical domain by using forward simulatability [8] originally introduced in natural language processing. We first start by formalizing simulatability, analyzing its limits in the medical domain, and proposing mitigation measures to overcome these limits. Forward simulatability serves as a key metric for assessing the performance and effectiveness of XAI models as it allows to measure the ability of human users, in particular clinicians, to simulate and understand the decision-making process of machine learning models. We then present three novel XAI models built over an existing MLBP, namely, the ML-RO model [10]. These XAI models will be used to experiment the XAI evaluation procedure based on forward simulatability.
Evaluating Explanations with Simulatability in the Medical Domain
Forward simulatability, as described by Hase et al. (2020) [8], represents a strategy employed to assess the effectiveness of explanation methods for decisions made by MLBPs (see Fig. 1). The assumption is an XAI model over an MLBP is good for a user if it enables the user to replicate the decisions of the MLBP over selected samples. Therefore, an explanation model for an MLBP is considered superior to another if it prompts the individual to make more decisions aligning with the MLBP’s intended outcomes. The forward simulatability approach is an objective and quantifiable measure to evaluate the quality and effectiveness of explanation methods. By comparing the decision-making alignment between individuals exposed to different explanation models, valuable insights can be gained regarding the suitability and reliability of these models in guiding decision-making processes.
To evaluate the efficacy of our XAI approach, we establish the forward simulability score (FSS) with respect to the specific MLBP under consideration as follows:
In the provided formula, D corresponds to the set of individual observations, f(x) represents the output decision made by the clinical expert, and m(x) signifies the decision output of the specific MLBP, for which explanations are being provided and
Applying the forward simulatability score in the medical domain presents unique challenges compared to its original proposal in the natural language processing setting [8]. Furthermore, these challenges are closely interconnected. In the following paragraphs, we analyze the challenges and propose measures to mitigate them.
The first challenge is that clinical decisions are inherently complex and time-consuming. If clinicians were exposed to all clinical variables during the decision-making process, it could significantly slow down their workflow. Given the characteristics of our MLBP [10], this aspect is mitigated in our work. Indeed, our system is based on dividing features into homogeneous groups that play a role in the decision-making process both at the group level and at the individual feature level. This enables us to offer evaluators a set of structured feature groups that closely resemble the information available to clinicians in a traditional medical record, reducing their time consumption.
The second challenge is even more subtle as simulatability can boost some explainability models only because these models expose features that are considered already relevant in specific clinical algorithms. Indeed, clinical algorithms [24,25,26] are key, well-established tools for medical decision-making and may represent a problem for evaluating explanations with simulatability. These algorithms are collections of rules generally organized in decision trees or flow charts where nodes are features. For this reason, it may happen that if the explanation proposes features that are in the clinical algorithm, clinicians may make decisions according to their clinical algorithm. To mitigate the bias of clinical algorithms in evaluating explainability methods, we propose to report on cases where the machine learning algorithm is in contrast with the clinical algorithm, where available. This may help to better understand the intrinsic value of explainability methods.
The last challenge is that, in some cases, the clinical task is hard, and cause-effect relations are unknown. Then, the MLBS may behave better than clinicians in finding the right answer or outcome, just because it can leverage a better pattern recognition system which has learned its model from a large amount of data. In this case, FSS can be low, but this does not necessarily imply that the explanation method is not good. To overcome this issue, the applicability of FSS should be carefully judged, and clinical decision tasks should be selected accordingly.
Machine Learning Predictors and Explanation Methods
In this paper, we align with the principles described in [8]. Specifically, we leverage a tabular dataset to investigate the significance of features within the decision-making process. Furthermore, we utilize information pertaining to feature groups, similar to the approach proposed in [27]. In the following section, we provide a concise explanation of our multiple kernel approach based on feature groups.
Group Features and Random Optimization
The decision function in the linear SVM is as follows:
where w is called the weight vector. This vector represents the importance of each feature in the decision-making process. The relevance of the feature \(w_i\) can be computed using the absolute value of \(w_i\), for each feature \(x_i \in X_{input}\).
Our approach leverages a multi-kernel learning strategy combined with random optimization to identify the optimal weights for the classification task, similar to what has been done in [27].
In the multi-kernel SVM algorithm decision function:
a Kernel function K can be utilized to map the input into high-dimensional feature space.
The weights assigned to the groups of input vector \(X_{input_g}\) with a \(\beta \) vectors can be computed as below:
in which \(X_{input_g}\) is the set of groups of features and \(\beta \) is defined as follows:
where \(\alpha _{g(i)}\) is the weight associated to the group i of the features.
During the learning phase, we perform random optimization (RO) iterations to find the best group weights configuration for the task. In each RO, the iteration starts from a random set of weights \(\alpha _{start}\) = [\(\alpha _0\), ..., \(\alpha _{|G|}\)]. We compute a perturbation of the \(\alpha \) vector and evaluate the model with the new \(\alpha _t\). We perform a perturbation by the value \(k_i\) for each \(\alpha _i\) in which each \(k_i\) is sampled from a uniform distribution between \(-lr\) and lr. At each time step \(t+1\), the new \(\alpha ^{t+1}\) vector is [\(\alpha _0^{t-1} + k_0\), ..., \(\alpha _{|G|}^{t-1} + k_{|G|}\)]. If the sum between \(\alpha _i^{t-1} + k_i\) is greater than 1, we set the new \(\alpha _i^{t+1}\) value to 1; otherwise, if the value is lower than 0, we set its value to 0. At each iteration, we transform the data by kernel function computation, and then we use transformed data to evaluate our model with the new \(\alpha ^{t+1}\) vector. We accept the new point only if it improves the average performance of the previous point over the 3-fold cross-validation on 80% of data which constitute the training set randomly chosen during the experiment definition phase, The remaining 20% of the data is allocated to the test set. Otherwise, we continue to find a better perturbation of \(\alpha ^t\).
The final value for the feature importance for each group is calculated as the min_max normalized value of the following expression:
where \(w_jx_j\) product is the importance value of individual features.
Explainability Methods
One of the most effective techniques for implementing an XAI approach is feature importance estimation [8]. In this study, we introduce a novel method for estimating feature importance that considers not only the significance of individual features but also the weights assigned to different feature groups within our MLBP.
To implement this method, we have proposed three different configurations of our XAI model: most important group features (MIGF), top K features by importance (TOPK), and features for groups (F4G). All three explanations are local explanation methods, as they are tailored to each individual patient record.
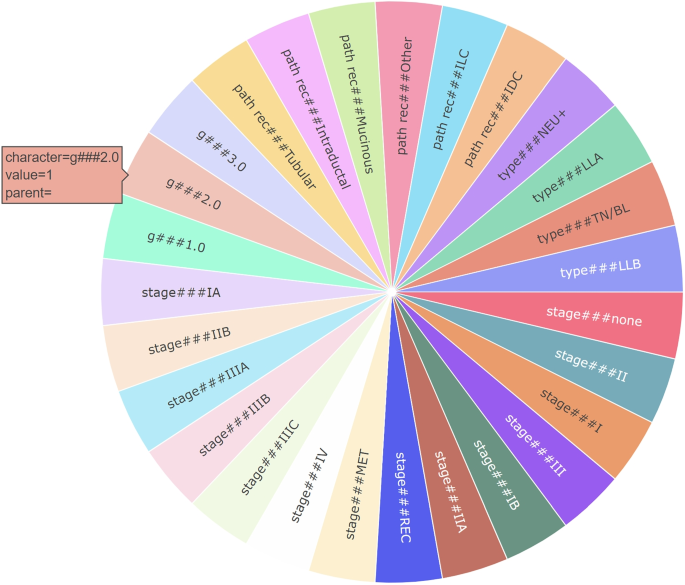
In the MIGF approach, we showcase all the features belonging to the most relevant group for the decision made, along with their weights within that group (see Fig. 2). This approach aims to focus clinicians’ attention only on the group that our MLBP deems to be the most important in the decision-making process for that specific patient. In conclusion, MIGF highlights the prominent aspect in the decision, but does not provide information regarding other groups of variables present in the model.
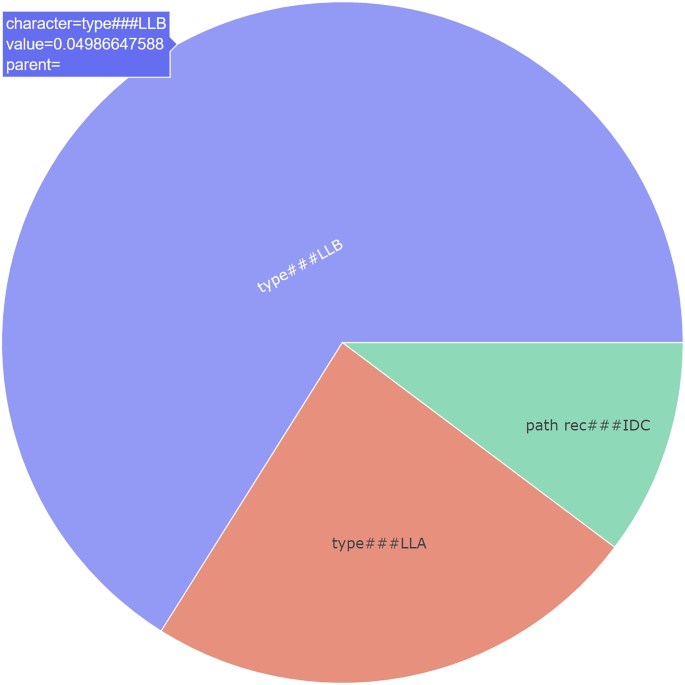
In the TOPK method, we present a visualization of the values and importance of the top K features, ordered by their significance in the decision, irrespective of their groups (see Fig. 3). By using this method, users are provided with the top K features that are most relevant to the prediction for a specific example. The visualization lets users identify the most critical features influencing the model’s decision, regardless of their parent group categories. The objective of this approach is to enhance the clarity and understanding of the model’s decision-making process by highlighting the level of importance associated with each feature in the decision. This implementation follows the standard approach for feature importance analysis.
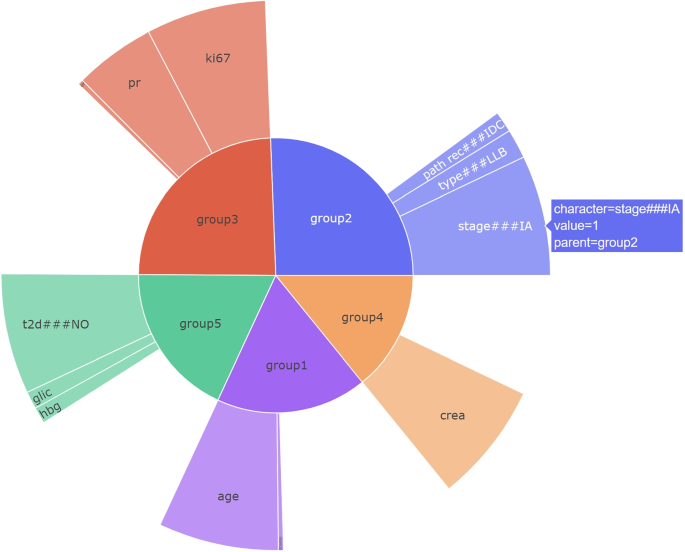
In the F4G method, we present the groups based on their importance in the decision, which is calculated similarly to the MIGF approach. Additionally, we display the features within each group, indicating their importance in the decision. This method provides a comprehensive view of the importance of both groups and individual features within the decision-making process (see Fig. 4). In this case, healthcare professionals can leverage both the importance of the groups and the importance of the corresponding features to understand how the MLBP system reached the final decision. By considering the significance of the groups and their constituent features, clinicians can gain insights into the decision-making process of the MLBP and evaluate the factors contributing to the final outcome. This comprehensive understanding enables them to make informed assessments and interpretations of the MLBP’s decisions in the context of patient care.
Challenging Clinical Datasets
To experiment with the FSS over our proposed explainability methods, we used three different datasets to study a variety of different clinical situations. The three datasets are venous thromboembolism event detection, breast cancer, and migraine. Each dataset is organized into several feature groups on which our explainability approach is based. In Table 1, the size of the dataset, the number of feature groups, and, if any, the reference medical prediction score are provided.
VTE Dataset
The VTE dataset aims to be the base to study the onset of venous thromboembolism (VTE)in cancer patients treated with chemotherapy [10]. The VTE dataset was obtained through the collaboration between the Policlinico Tor Vergata Medical Oncology Unit (PTV) and the BioBIM (InterInstitutional Multidisciplinary Biobank, IRCCS San Raffaele Pisana). It consists of 1179 ambulatory cancer patients with primary or relapsing/recurrent solid cancers. The VTE dataset consists of 11 groups of features comprising a total of 37 features (see Table 1). The event of interest is the occurrence of the venous thromboembolic event during cancer therapies.
The VTE dataset is extremely important for our study as it is strongly connected to a clinical algorithm: the Koranha score (KS), a nearly standard clinical algorithm to estimate the risk of VTE in cancer patients. Hence, the KS can trigger the decisions of clinicians that are evaluating MLBPs with forward simulatability. As a mitigation measure, we presented clinicians only with samples where the MLBP is in disagreement with KS. This will filter out the bias of evaluating the ability of the explanation method to replicate the clinical algorithm.
Breast Cancer Dataset
The breast cancer (BC) dataset was obtained through collaboration between the Policlinico Tor Vergata Medical Oncology Unit (PTV) and the BioBIM (InterInstitutional Multidisciplinary Biobank, IRCCS San Raffaele Pisana), and consists of 454 patients diagnosed with breast cancer. For the present analysis, only patients for whom prognostic and pre-treatment biochemical factors were available were selected. The dataset was split into a training set consisting of 318 patients and a test set comprising 136 patients. Its features are organized into five groups. The target event is determining the progression of the disease in BC patients [28].
The BC dataset is not related to any clinical algorithm. Still, it represents a specific clinical prediction whose cause-effect relations between independent variables and dependent variables are not completely clear. Apart from the specific clinical and biochemical features of breast cancer of known prognostic and predictive value, the use of routinely available biochemical parameters and their interaction with the aforementioned specific features and impact on disease progression are not fully known. Thus, one of the possible successes of an explanation method for an MLBP can be the ability to show possible cause-effect relations among all the possible ad hoc relations used to take the final decision over the prediction.
Migraine Dataset
Migraine dataset [29] consists of 739 patients recruited by the Headache and Pain Unit of the Department of Neurological, Motor and Sensorial Sciences and the InterInstitutional Multidisciplinary Biobank (BioBIM) of the IRCCS San Raffaele Pisana, Rome, Italy, starting from January 2008. Its features are organized into six groups, and the analyzed event is medication overuse in patients. The target event is drug overuse.
The migraine dataset represents a specific clinical prediction whose cause-effect relations between independent variables and dependent variables are not completely clear.
Results and Discussion
The ML-RO system is a good model on which to test our XAI evaluation strategy based on the FSS. Indeed, ML-RO, which is the MLBP whose explanation models are evaluated in this study, outperforms two other base models—a decision tree (DT) and a plain SVM—in the three datasets (Table 2).
It is extremely important that ML-RO significantly outperforms the transparent decision tree (DT) learning model. Indeed, DT would have an FSS of 1 as the decision tree is a decision algorithm that is in itself an explanation of the decision process. Hence, clinicians can easily replicate predictions of the decision tree. However, DT has an F-score of 17% lower than ML-RO for BC, 1.3% for VTE, and 22% for migraine. It is also important to have a more robust model that can be explained. Here, ML-RO can outperform DT and, at the same time, important factors can be extracted in the algorithm decision-making.
Moreover, ML-RO outperforms plain SVM, and the F4G explanation model is possible only using ML-RO. ML-RO improves SVM by using a multiple kernel approach and an optimization technique—random optimization—that optimizes the relative weights of the groups of features. Besides that, the fact that ML-RO outperforms SVM in all tasks is crucial as ML-RO also offers the possibility to utilize group features for the explanation in F4G, which is not possible for the general SVM approach. It is essential to offer and evaluate an explanation model for a more obscure but fairly accurate MLBP as better-performing models are usually harder to explain [30, 31].
Having a good MLBP and three XAI models, we can test our evaluation strategy of XAI models. Then, we involved two clinicians for BC and VTE explanation experiments and one in the evaluation of Migraine. The clinical experts analyzed ten cases for each dataset, and for each of them, they provided human predictions based on raw data and human predictions based on the three explanation models (MIGF, TOPK, F4G). The three clinicians, utilizing all three explainability methodologies together to perform forward simulatability, emphasized that only F4G could be used independently, while MIGF and TOPK, although providing useful information for certain aspects, were not sufficiently informative unless combined with the other two methods, particularly F4G.
The first result of the clinicians is that F4G is the most appropriate method for explaining ML-RO. In general, the introduction of feature groups in both the MLBP and the explainability method has brought significant advantages.
The second result is that the FSS is useful to understand the validity of a specific XAI model over an MLPS. Indeed, FFS suggests that the explanation method F4G for the ML-RO is effective for two datasets out of the three, and it shows the limits of the learned model for one dataset. In fact, the FSS improves by 10% after using the explanation F4G for BCand 5% on VTE (see Table 3). The explanation model is expressive enough to allow clinicians to make the correct decisions.
Finally, the last result is that FSS can highlight artifacts in data that incorrectly drive decisions of MLPS. There is also another interesting result provided by the Migraine dataset (see Table 3). This result seems to reduce the importance of the explanation model. There is a drop in FFS after the explanation. This may appear as bad news for the explanation model, but, on the contrary, it has a good effect on the explanation model. The explanation model points out some problems in the ML-RO decision model. Decisions were indeed made for the wrong reason—the artifact—by looking at an implausible feature. To cope with missing feature values, the migraine dataset has been augmented with classical feature-filling methods, that is, filling missing numerical feature values with the mean value of that feature in the dataset. In this case, clinicians found that the system has replaced some empty values with an implausible value, e.g., a value for the feature menarche for a male subject. Hence, in this case, the explainability model helped to understand that MLBP takes the right decision for the wrong reason, as the MLBP accuracy on the selected cases is 1.
Results of the experiments show the effectiveness of the strategy based on forward simulatability that we propose to evaluate XAI methods in the medical domain. Indeed, it is possible to evaluate XAI models with FSS. For the proposed XAI model, the increase in FSS is consistently positive, indicating that FSS can be used as a method to rank different XAI models.
Conclusions
Explaining is crucial to make machine learning-based predictors (MLBPs) acceptable in medical practice and a way to evaluate explanation methods is needed [1]. In this paper, we proposed a methodology to evaluate explanation methods for MLBPs in the clinical setting based on forward simulatability. We highlighted the pitfalls of using this method in the clinical setting. Our study aims to introduce a standard practice for evaluating MLBPs along with their explanation methods, with the goal of promoting transparency, trustworthiness, and interpretability in the application of machine learning algorithms within the medical field. By establishing a rigorous evaluation framework, we seek to provide healthcare professionals with reliable tools for assessing the performance and reliability of MLBPs, ultimately enhancing their adoption and integration into clinical practice.
Despite the promising results obtained in setting out a procedure to evaluate XAI methods in medicine and the experiments highlighting the utility of our approach in improving the performance of our MLBP, there are certain potential limitations and future developments that need to be addressed. First, the XAI evaluation procedure should be assessed in comparison with questionnaires aiming to assess whether XAIs are increasing the level of trust in MLBPs. Second, the evaluation procedure should be tested on more clinicians. However, to obtain this large-scale analysis involving many clinicians, the related MLBP should be integrated in a medical device. This is a long procedure requiring MLBPs to have already gained the trust of clinicians. Finally, the European Artificial Intelligence ActFootnote 1 has opened new challenges. Finally, the issue that should be investigated is whether our XAI evaluation procedure can show the compliance of XAI-empowered MLBPs and the AI Act.
Data Availability
The data that support the findings of this study are not openly accessible due to reasons of sensitivity but are available from the corresponding author upon reasonable request. Data are located in controlled access data storage at BIOBIM, University of Rome Tor Vergata, and at the Headache and Pain Unit of IRCSS San Raffaele Roma.
References
May M. Eight ways machine learning is assisting medicine. Nat Med. 2021;27(1):2–3. Number: 1 Publisher: Nature Publishing Group. https://doi.org/10.1038/s41591-020-01197-2.
Lu SC, Swisher CL, Chung C, Jaffray D, Sidey-Gibbons C. On the importance of interpretable machine learning predictions to inform clinical decision making in oncology. Front Oncol. 2023;13:1129380. https://doi.org/10.3389/fonc.2023.1129380.
Amann J, Blasimme A, Vayena E, Frey D, Madai VI. Explainability for artificial intelligence in healthcare: a multidisciplinary perspective. BMC Med Inform Decis Mak. 2020. https://doi.org/10.1186/s12911-020-01332-6.
Banegas-Luna AJ, Peña-García J, Iftene A, Guadagni F, Ferroni P, Scarpato N, et al. Towards the interpretability of machine learning predictions for medical applications targeting personalised therapies: a cancer case survey. Int J Mol Sci. 2021;22(9):4394. https://doi.org/10.3390/ijms22094394.
Sokol K, Flach P. Explainability fact sheets: a framework for systematic assessment of explainable approaches. In: Proceedings of the 2020 Conference on Fairness, Accountability, and Transparency. FAT* ’20. New York, NY, USA: Association for Computing Machinery; 2020. pp. 56–67.
Coroama L, Groza A. Evaluation metrics in explainable artificial intelligence (XAI). In: Guarda T, Portela F, Augusto MF, editors. Advanced Research in Technologies, Information, Innovation and Sustainability. Cham: Springer Nature Switzerland; 2022. pp. 401–13.
Belle V, Papantonis I. Principles and practice of explainable machine learning. Front Big Data. 2021;4:688969. https://doi.org/10.3389/fdata.2021.688969.
Hase P, Bansal M. Evaluating explainable AI: which algorithmic explanations help users predict model behavior? In: Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics. Online: Association for Computational Linguistics; 2020. pp. 5540–52. https://aclanthology.org/2020.acl-main.491.
Bansal G, Nushi B, Kamar E, Lasecki WS, Weld DS, Horvitz E. Beyond accuracy: the role of mental models in human-AI team performance. Proceedings of the AAAI Conference on Human Computation and Crowdsourcing. 2019;7(1):2–11. https://doi.org/10.1609/hcomp.v7i1.5285.
Ferroni P, Zanzotto FM, Scarpato N, Riondino S, Nanni U, Roselli M, et al. Risk assessment for venous thromboembolism in chemotherapy-treated ambulatory cancer patients. Med Decis Making. 2016;37(2):234–42. https://doi.org/10.1177/0272989X16662654.
Gilpin LH, Bau D, Yuan BZ, Bajwa A, Specter M, Kagal L. Explaining explanations: an overview of interpretability of machine learning. In: 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA), Turin, Italy. 2018. pp. 80–9. https://doi.org/10.1109/DSAA.2018.00018.
Burkart N, Huber MF. A survey on the explainability of supervised machine learning. J Artif Intell Res. 2021;70:245–317. https://doi.org/10.1613/jair.1.12228.
Arrieta AB, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado A, et al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion. 2020;58:82–115.
Linardatos P, Papastefanopoulos V, Kotsiantis S. Explainable AI: a review of machine learning interpretability methods. Entropy. 2020;23(1):18.
Ali S, Abuhmed T, El-Sappagh S, Muhammad K, Alonso-Moral JM, Confalonieri R, et al. Explainable artificial intelligence (XAI): what we know and what is left to attain trustworthy artificial intelligence. Inf Fusion. 2023. https://doi.org/10.1016/j.inffus.2023.101805.
Zhou J, Gandomi AH, Chen F, Holzinger A. Evaluating the quality of machine learning explanations: a survey on methods and metrics. Electronics. 2021;10(5):593. Number: 5 Publisher: Multidisciplinary Digital Publishing Institute. https://doi.org/10.3390/electronics10050593.
Moradi M, Samwald M. Post-hoc explanation of black-box classifiers using confident itemsets. Expert Syst Appl. 2021;165:113941. https://doi.org/10.1016/j.eswa.2020.113941.
Quinlan JR. C4. 5: programs for machine learning. Elsevier; 2014.
Lipton ZC. The mythos of model interpretability: in machine learning, the concept of interpretability is both important and slippery. Queue. 2018;16(3):31–57.
Borys K, Schmitt YA, Nauta M, Seifert C, Krämer N, Friedrich CM, et al. Explainable AI in medical imaging: an overview for clinical practitioners - Saliency-based XAI approaches. Eur J Radiol. 2023;162:110787. https://doi.org/10.1016/j.ejrad.2023.110787.
De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24(9):1342–50.
Ribeiro MT, Singh S, Guestrin C. “Why should I trust you?”: explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. KDD ’16. New York, NY, USA: Association for Computing Machinery; 2016. pp. 1135–44.
Ribeiro MT, Singh S, Guestrin C. Anchors: high-precision model-agnostic explanations. In: Proceedings of the AAAI Conference on Artificial Intelligence. 2018. https://doi.org/10.1609/aaai.v32i1.11491.
Lindström J, Tuomilehto J. The diabetes risk score: a practical tool to predict type 2 diabetes risk. Diabetes Care. 2003;26(3):725–31. https://doi.org/10.2337/diacare.26.3.725.
Khorana AA, Kuderer NM, Culakova E, Lyman GH, Francis CW. Development and validation of a predictive model for chemotherapy-associated thrombosis. Blood. 2008;111(10):4902–7.
Nunez TC, Voskresensky IV, Dossett LA, Shinall R, Dutton WD, Cotton BA. Early prediction of massive transfusion in trauma: simple as ABC (Assessment of Blood Consumption)? The Journal of Trauma: Injury, Infection, and Critical Care. 2009;66(2):346–52. https://doi.org/10.1097/ta.0b013e3181961c35.
Gönen M, Alpaydın E. Multiple kernel learning algorithms. J Mach Learn Res. 2011;12:2211–68.
Ferroni P, Zanzotto FM, Riondino S, Scarpato N, Guadagni F, Roselli M. Breast cancer prognosis using a machine learning approach. Cancers. 2019;11:1–9. https://doi.org/10.3390/cancers11030328.
Ferroni P, Zanzotto FM, Scarpato N, Spila A, Fofi L, Egeo G, et al. Machine learning approach to predict medication overuse in migraine patients. Comput Struct Biotechnol J. 2020;18:1487–96. https://doi.org/10.1016/j.csbj.2020.06.006.
Wu L, Huang R, Tetko IV, Xia Z, Xu J, Tong W. Trade-off predictivity and explainability for machine-learning powered predictive toxicology: an in-depth investigation with Tox21 data sets. Chem Res Toxicol. 2021;34(2):541–9.
Nauta M, Trienes J, Pathak S, Nguyen E, Peters M, Schmitt Y, et al. From anecdotal evidence to quantitative evaluation methods: a systematic review on evaluating explainable AI. ACM Comput Surv. 2023;55(13). https://doi.org/10.1145/3583558.
Funding
This work was partially funded by grants from the European Project Horizon 2020 SC1-BHC-02-2019 (REVERT, ID:848098) and the European Project Horizon 2020 KATY (grant agreement No 101017453). The funding sources had no involvement in the study design; in the collection, analysis, and interpretation of data; in the report’s writing; and in the decision to submit the article for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Legal and Ethical Aspects
In this paper, we exploited three distinct datasets obtained from reputable research institutions (BIOBIM, University of Rome Tor Vergata, IRCSS San Raffaele Roma). The data collection, compilation, and storage procedures adhered to the current regulations, including the General Data Protection Regulation (GDPR), and were conducted in accordance with the ethical principles outlined in the Declaration of Helsinki.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Scarpato, N., Nourbakhsh, A., Ferroni, P. et al. Evaluating Explainable Machine Learning Models for Clinicians. Cogn Comput (2024). https://doi.org/10.1007/s12559-024-10297-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12559-024-10297-x