Abstract
Atrophy measured on structural magnetic resonance imaging (sMRI) is a powerful biomarker of the stage and intensity of the neurodegenerative aspect of Alzheimer's disease (AD) pathology. In this review, we will discuss the role of sMRI as an AD biomarker by summarizing (a) the most commonly used methods to extract information from sMRI images, (b) the different roles in which sMRI can be used as an AD biomarker, and (c) comparisons of sMRI with other major AD biomarkers.
Similar content being viewed by others
Pathological cascade and structural magnetic resonance imaging
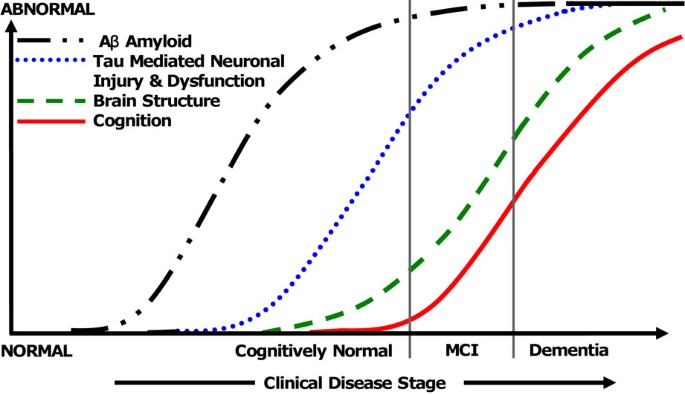
Alzheimer's disease (AD) is a multifaceted disease in which cumulative pathological brain insults result in progressive cognitive decline that ultimately leads to dementia. Amyloid plaques, neurofibrillary tangles (NFTs), neurodegeneration, and inflammation are the well-established pathological hallmarks of AD. A plausible model for the development of AD posits that amyloid deposition occurs early in the process but by itself does not directly cause clinical symptoms [1, 2]. Neuronal and synaptic losses appear to be key determinants of cognitive impairment in AD [3, 4]. If neuronal loss leads to cerebral atrophy (as is likely), then it can be expected that cognitive decline and atrophy will be closely associated. On the basis of this evidence, it has been hypothesized that AD pathological cascade is a two-stage process in which amyloidosis and neuronal pathology (tauopathy, neuronal injury, and neurodegeneration) are largely sequential rather than simultaneous processes [1, 5, 6]. There is also sufficient literature to support the fact that atrophy of the brain structures or neurodegeneration is the most proximate substrate of cognitive impairment in AD [2, 7–9]. This hypothesis of a sequential model was proposed by Jack and colleagues [6] on the basis of biomarker data and is adapted and illustrated in Figure 1. Owing to the close relationship between neurodegeneration and cognition (as illustrated in Figure 1), atrophy measured on structural magnetic resonance imaging (sMRI) is a powerful AD biomarker.
Proposed Alzheimer's disease pathological cascade based on biomarkers. MCI, mild cognitive impairment. Modified and reproduced with permission from [6].
sMRI measures brain morphometry and therefore can capture gray matter atrophy related to the loss of neurons, synapses, and dendritic de-arborization that occurs on a microscopic level in AD; white matter atrophy related to the loss of structural integrity of white matter tracts, presumably resulting from demyelination and dying back of axonal processes; and ex vacuo expansion of cerebrospinal fluid (CSF) spaces. Since there is a significant negative correlation between NFT density and neuronal counts [10], sMRI indirectly reflects NFT density. It has been shown that neuronal loss correlates with but exceeds NFT density in AD and is related directly to impaired cognitive function [10]. Neuronal loss also correlates with Braak NFT stage and quantitative NFT burden, validating sMRI as an AD biomarker [11–13]. This review provides a summary of the role of sMRI as an AD biomarker. First, we begin with the most commonly used methods to extract information from sMRI images, then we discuss the different roles in which sMRI can be used as a biomarker in AD, and finally we compare the performance of sMRI to that of other major AD biomarkers.
Extracting information from structural magnetic resonance imaging
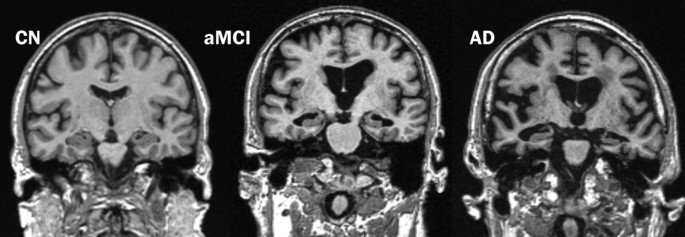
Given the large amount of data present in a three-dimensional (3D) sMRI scan, several different methods are employed to condense atrophy information in each patient's scan or assess atrophy over multiple scans of the same individual. The pattern of neurodegeneration seen using sMRI is similar to the progression of neurofibrillary pathology as described by Braak and Braak [14]. The disease usually begins and is ultimately most severe in the medial temporal lobe, particularly the entorhinal cortex and hippocampus. Later (that is, when subjects are in the clinical mild cognitive impairment [MCI] phase), the disease spreads to the basal temporal lobe and paralimbic cortical areas such as the posterior cingulate gyrus and precuneus. The onset of dementia is due to the spread of degenerative atrophy to multimodal association neocortices. Basal forebrain and the dorsal pontomesencephalic areas are also involved. However, unusual variants that do not follow this particular pattern are increasingly recognized. Furthermore, other limbic lobe structures such as posterior cingulate seem to be involved early and consistently in AD. Figure 2 shows typical MRI scans in cognitively normal (CN) subjects and in patients with MCI or AD. As can been seen in the figure, there is increasing medial temporal atrophy (specifically, the hippocampus and ventricular enlargement) in MCI and AD when compared with CN. Here, we present a brief survey of methods to extract or visualize this information (or both) from 3D sMRI scans of cross-sectional and longitudinal studies.
Cross-sectional methods
When changes in different individuals are measured cross-sectionally, the most widely used summary measures from sMRI are the following:
1. Visual assessment of scans
Often, visual assessment of the degree of atrophy in the medial temporal lobe is used as a metric to measure disease [15, 16]. Visual assessment offers a fast and efficient way to assess MRI scans but does not capture the fine incremental grades of atrophy.
2. Quantitative region of interest-based techniques or volumetry
Volumetry is the most common cross-sectional quantitative metric used in AD. Although traditionally manual tracing of volumes was used, the increase in computational power has led to the development of automated techniques.
2a. Manual tracing
Tracing and quantifying the volume of medial temporal lobe structures (for example, the hippocampus or entorhinal cortex) or posterior cingulate have been traditionally employed in AD and provide an accurate quantitative measure of atrophy [17]. However, manual measurements can be tedious and time-consuming.
2b. Automated and semi-automated techniques
In the recent past, methods have been proposed to automatically parcellate gray matter density or cortical surfaces into regions of interest. These cortical surfaces are used to compute global as well as a regional cortical thickness (that is, combined thickness of the layers in the cerebral cortex). Because automated and semi-automated techniques do not require significant manual intervention, they are extremely useful for large-scale studies.
An advantage of volumetry, such as measuring the hippocampus, is that the measurements describe a known anatomic structure that (in the case of the hippo-campus) is closely related to the pathological expression of the disease and is also functionally related to one of the cardinal early clinical symptoms - memory impairment. However, the disadvantage of using a single region of interest to consolidate 3D information as a disease metric is that it is spatially limited and does not make use of all of the available information in a 3D sMRI.
3. Quantitative voxel-based
These methods assess atrophy over the entire 3D sMRI scan.
3a. Voxel-based analytic techniques
Methods such as voxel-based morphometry (VBM) [18] have been developed to provide a powerful way to test for group-wise comparisons between cross-sectional sMRI scans of diseased group versus normal controls. The typical atrophy patterns seen in subjects with AD or MCI are similar to those of the Braak neurofibrillary staging described above. Although VBM enables visualization of the pattern of neurodegeneration due to disease, the statistical testing portion of VBM is designed only to test for group-wise differences between two groups of subjects and cannot provide a summary measure for each subject, and this makes it inapplicable to diagnosis in individual subjects.
3b. Automated individual subject diagnosis
Several investigators have recently turned their attention to multivariate analysis and machine learning-based algorithms that use the entire 3D sMRI data to form a disease model against which individual subjects may be compared. These scores typically are computed for each new incoming scan (that is, test scan) on the basis of the degree and the pattern of atrophy in comparison with the scans of a large database of well-characterized AD and cognitively normal subjects [19–22].
Longitudinal methods
Because accelerating tissue loss is a hallmark of neuro-degenerative disease, serial sMRI scans often are analyzed to measure disease progression. Even though cross-sectional measures can be employed to obtain a summary measure from sMRI at every time point, these measures have unnecessary variability due to inherent noise associated with each individual measurement. Therefore, specific techniques have been developed to extract tissue loss information from serial sMRI scans. In these techniques, all pairs of sMRI scans are registered to each other and brain loss between scans is quantified and this reduces the variability.
Global atrophy quantification
One of the earliest methods developed to quantify the global percentage change in brain volume between two scans was boundary shift integral (BSI) [23]. BSI determines the total volume through which the surface of the brain has moved between scans acquired at two time points (that is, the brain volume decreases and the volume of the ventricles increases). One of the most sensitive global measures for measuring the rates of brain atrophy is the ventricular change measure using BSI [24]. This is because the ventricular boundary on sMRI (T1-weighted images) provides a good contrast for the delineation of the ventricular surface with more accuracy when compared with brain volume and hippocampal volume.
Tensor-based morphometry
Unlike BSI, which analyzes only spatial shift in the brain surfaces, TBM provides a 3D profile of voxel-level brain degeneration. Here, the term TBM is used to describe 3D voxel-based methods that can be employed to observe how the disease progresses in the brain as a result of the underlying pathological changes [25, 26].
Role of structural magnetic resonance imaging in Alzheimer's disease and mild cognitive impairment
In this section, we will briefly discuss the different roles in which sMRI can be employed as an AD biomarker. When MCI involves primarily memory complaints and deficits, it is often considered a prodromal stage of AD. Here, we will also discuss the role of sMRI in MCI in addition to AD.
1. Early diagnosis of Alzheimer's disease and mild cognitive impairment
The typical reductions of hippocampal volume in MCI with an average Mini-Mental State Exam (MMSE) score of 25 are 10% to 15% and in AD with an average MMSE score of 20 are 20% to 25% [27]. Measuring these significant reductions (due to AD) in the medial temporal lobe can be extremely useful for early diagnosis of AD and MCI. At present, diagnostic criteria for AD are based on the criteria in the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV), which are based primarily on clinical and psychometric assessment and do not use quantitative atrophy information available in sMRI scans. However, there is a proposal to add reliable biomarkers to the diagnostic criteria [28]. One of the suggested features is the volume loss of medial temporal structures since measures of sMRI atrophy have accuracies of 70% to 90% in AD and 50% to 70% in amnestic MCI in distinguishing them from age-matched controls [28]. All of the above-mentioned cross-sectional methods, except 3a, can be used as diagnostic metrics for AD and MCI.
2. Predicting the risk of progression in mild cognitive impairment and cognitively normal
Although there is considerable variability of progression rates in MCI to AD, it has been observed that an average of about 10% to 15% of subjects with MCI, specifically of the amnestic type, annually progress to AD [29]. Because pathological changes occur before the onset of clinical symptoms, biomarkers can aid in the prediction of risk of progression in MCI and CN. A recent meta-analysis showed that hippocampal volume can detect an average of approximately 73% of MCI subjects who progress to AD [30]. Several studies using both cross-sectional methods 1 and 2 above have shown that atrophy seen on MRI can predict the risk of progression to AD with good accuracy.
3. Evaluating disease progression
Charting structural changes in the brain over time is important in monitoring the progression of the disease [31]. Tracking the disease progression is especially important in patients with MCI and cognitively normal subjects since atrophy rates can predict subsequent clinical progression in both groups. The metrics that are most often used for evaluating or tracking disease progression are increase in ventricular volume and decrease in brain volume over time. These measures are more sensitive than cross-sectional measures in capturing changes over time since all scans of the same subject are registered together to reduce inter-scan variability.
4. Measuring the efficacy of therapeutics
Several investigators have shown that the lower variance in the serial sMRI measurements compared with clinical measures of cognition and function could permit clinical trials to be performed with smaller sample sizes than would be possible using traditional clinical instruments [32–34]. At present, AD biomarkers have not yet been validated as surrogate endpoints for regulatory purposes and therefore cannot be used as the primary indicators of efficacy. However, the impact of interventions on these biomarkers has been evaluated in a few trials and was found to be potentially useful in capturing the pharmacodynamic effects. The efficacy of donepezil, a cholinesterase inhibitor, was evaluated using serial sMRI [35, 36] and was found to possibly be neuro-protective in nature since there was some evidence for decreased disease progression on the basis of sMRI trophy. In a different study, it was observed that subjects immunized with Aβ antibody responders had a more rapid volume loss than placebo patients during a phase IIa immunotherapy trial that was prematurely terminated owing to meningoencephalitis in a subset of patients [37]. In addition to evaluating therapeutic efficacy, atrophy on sMRI can be used to select at-risk MCI subjects for clinical trials. While longitudinal methods are useful for testing efficacy of therapeutics, cross-sectional methods are most suited for sample enrichment.
5. Screening in clinical trials
MRI is routinely used at two stages in clinical trials. The first is screening at baseline for inclusion/exclusion. This includes identifying subjects with imaging evidence of conditions that are exclusionary (for example, hemispheric infarction or prior evidence of cerebral hemorrhage). Also, anti-amyloid trials commonly will exclude subjects with micro-hemorrhages that exceed a specified number. Either long echo time gradient echo or susceptibility-weighted imaging sequences are used for micro-hemorrhage identification. MRI is also used for safety screening during the study. Conditions that are of interest are evidence of new micro-hemorrhage and vasogenic edema. FLAIR (fluid-attenuated inversion recovery) and diffusion imaging are used to identify the latter condition.
6. Differential diagnosis of dementia subtypes
Given that pathology does not always map onto the clinical expression of the disease and has considerable clinical heterogeneity, biomarkers such as sMRI can aid in the differential diagnosis of dementia types. The absence of significant medial temporal lobe atrophy in dementia with Lewy bodies [38] and vascular dementia [39], significant frontal lobe atrophy in behavioral variant fronto-temporal dementia [40], or pronounced asymmetrical temporal lobe atrophy in semantic dementia [41] can be used to separate these non-AD dementias from AD. Diffusion imaging and FLAIR are useful in identifying both cerebrovascular disease and prion disease. MRI is useful in identifying structural contributors to cognitive impairment such as hemorrhage or evidence of major head trauma. Differential diagnosis of dementias using sMRI will be particularly helpful when therapeutics become readily available.
7. Mechanistic inferences into the disease process
Using sMRI as an independent biomarker of neurodegeneration aides in understanding relationships between cognition and neurodegeneration in AD. This has led to insights into disease mechanisms in AD. In the model shown in Figure 1 from Jack and colleagues [6], the conclusion that neurodegeneration is more proximately associated with cognitive decline was derived from several sMRI studies.
Comparison of structural magnetic resonance imaging with other major Alzheimer's disease biomarkers
The major AD biomarkers that are typically considered for clinical trials and observational studies are CSF Aβ1-42, CSF t-tau, fluoro-deoxy-glucose positron emission tomography (FDG-PET), Pittsburgh compound B-PET (PIB-PET), and sMRI. In this section, we will compare sMRI with other major AD biomarkers by summarizing studies that have compared sMRI with each of these biomarkers in the same set of subjects.
Structural magnetic resonance imaging and cerebrospinal fluid
Low CSF Aβ1-42 levels reflect deposition of Aβ in plaques, high CSF t-tau reflects active axonal and neuronal damage, and high p-tau reflects phosphorylated-tau and has been postulated to more closely mirror NFT formation. Several CSF and sMRI studies have compared the diagnostic and prognostic accuracy of both and have attempted to characterize the associations between the two biomarkers in the same set of subjects. We have summarized these studies in Table 1. The majority of the studies have concluded that sMRI and CSF provide independent diagnostic information and that the combination provides better discrimination of AD than either one does alone [42–44]. It has also been shown that both biomarkers are good predictors of MCI progression to AD [45–47]. However, the associations between both of the biomarkers have not been consistent across studies. While some studies claim that there is an association between CSF biomarkers (specifically t- tau and p-tau) and sMRI [42, 46, 48–54], others have found no association between the two [45, 55–57]. This could be due mainly to the fact that measuring the biomarkers in different study populations (that is, at different stages of the disease) will provide different answers, and also there is a large variability in the methodologies used (that is, variability in the assays and sMRI measures ranging from visual assessment to automated diagnosis).
The earlier studies concentrated mainly on the associations between CSF and sMRI biomarkers, whereas the more recent ones have started investigating the association between these biomarkers and cognition. Studies published on the basis of the Alzheimer's Disease Neuroimaging Initiative (ADNI) data have shown that sMRI is more closely related to cognition than CSF biomarkers are [34, 43, 44, 47, 55], lending support to the model in Figure 1. As suggested by Wahlund and Blennow [48], CSF Aβ denotes a specific molecular pathway or etiology whereas CSF tau, p-tau, and sMRI may reflect the disease stage or intensity of AD. However, sMRI appears to be a more stable indicator of neuronal loss in comparison with the CSF measures. This may be due to the fact that brain volume quantification with sMRI has nothing analogous to daily turnover of a soluble protein measured using CSF.
Structural magnetic resonance imaging and FDG-PET
Decreased FDG-PET uptake (that is, hypo-metabolism on FDG-PET scans) reflects metabolic deficits due to synaptic dysfunction and (probably) tau-mediated neuronal injury. sMRI atrophy is seen mainly in the medial temporal lobes, whereas FDG uptake decreases are seen mainly in the posterior cingulate and parietal lobes. Studies that have investigated FDG and MRI in the same group of subjects are summarized in Table 2. Several studies have compared FDG and sMRI on the basis of diagnostic and prognostic accuracy in AD. FDG was found to provide slightly better discrimination than MRI in [58–62], and a couple of recent studies based on ADNI data found that the two have similar performance [44, 63] and have largely overlapping value for discrimination [44]. However, the question of complementary or overlapping information between FDG and sMRI remains to be investigated in a large group of subjects in a systematic fashion.
Structural magnetic resonance imaging and PIB-PET
Although there are several amyloid imaging PET tracers based on 11C and 18F, the tracer most studied in the field of AD is PIB [64], which we discuss here. PIB-PET scans measure the deposition of Aβ in the brain (amyloid load). Since the invention of PIB, there has been significant interest in investigating the effect of Aβ plaques as measured by PIB [64] on cognition and sMRI. In this section, we will discuss studies that have investigated both PIB and sMRI in the same group of subjects. These studies are summarized in Table 3. In CN, baseline PIB was not associated with longitudinal sMRI changes in the preceding years [65] but was strongly related to brain atrophy [66, 67] and future cognitive decline [66]. The majority of studies have found a correlation between baseline sMRI and PIB measures [68–70]. In addition, serial PIB and sMRI studies have found that longitudinal changes are much more pronounced on sMRI and that longitudinal change in PIB is minimal [1, 71]. All of this evidence has led to our understanding that Aβ deposition measured by PIB is an upstream process whereas neurodegeneration is a downstream process that is probably initiated by Aβ deposition and is more closely related to cognitive decline [1, 2].
Conclusions and future directions in structural magnetic resonance imaging
Given that the clinical assessment is unlikely to exactly match findings at autopsy in every subject, in vivo imaging measures (such as sMRI) that reflect disease stage and intensity would be extremely useful. The value added to clinical assessment by MRI is that it is an independent non-invasive measure of neuronal loss and thus provides a supplementary measure based only on anatomy; by contrast, clinical diagnosis is done on the basis of clinical examination and neuropsychological tests. Numerous studies now show that sMRI is a stable biomarker of AD progression. Publications on sMRI data from multicenter studies such as ADNI have also provided evidence that the combination of sMRI scans from multicenter studies is possible without much penalty [72]. In addition to being of diagnostic and prognostic value, sMRI can play multiple roles, as described in this review.
Three future directions still need to be thoroughly investigated. (a) The development of robust, validated, and automated techniques for extracting disease-specific information from cross-sectional and serial sMRIs needs to be investigated. (b) Because the majority of the studies discussed here were done on highly screened populations, it is important to validate the generalizability of sMRI as a biomarker in clinically based cohorts in which the presence of multiple pathologies and disorders is a norm rather than an exception. (c) How these sMRI measures can be integrated with other clinical measures, CSF, and PET biomarkers to be of clinical use needs to be investigated.
Abbreviations
- 3D:
-
three-dimensional
- AD:
-
Alzheimer's disease
- ADNI:
-
Alzheimer's Disease Neuroimaging Initiative
- BSI:
-
boundary shift integral
- CN:
-
cognitively normal
- CSF:
-
cerebrospinal fluid
- FDG:
-
fluoro-deoxy-glucose
- FDG-PET:
-
fluoro-deoxy-glucose positron emission tomography
- FLAIR:
-
fluid-attenuated inversion recovery
- MCI:
-
mild cognitive impairment
- MMSE:
-
Mini-Mental State Exam
- MRI:
-
magnetic resonance imaging
- NFT:
-
neurofibrillary tangle
- PET:
-
positron emission tomography
- PIB:
-
Pittsburgh compound B
- sMRI:
-
structural magnetic resonance imaging
- VBM:
-
voxel-based morphometry.
References
Jack CR, Lowe VJ, Weigand SD, Wiste HJ, Senjem ML, Knopman DS, Shiung MM, Gunter JL, Boeve BF, Kemp BJ, Weiner M, Petersen RC, Alzheimer's Disease Neuroimaging Initiative: Serial PIB and MRI in normal, mild cognitive impairment and Alzheimer's disease: implications for sequence of pathological events in Alzheimer's disease. Brain. 2009, 132: 1355-1365. 10.1093/brain/awp062.
Mormino EC, Kluth JT, Madison CM, Rabinovici GD, Baker SL, Miller BL, Koeppe RA, Mathis CA, Weiner MW, Jagust WJ, Alzheimer's Disease Neuroimaging Initiative: Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009, 132: 1310-1323. 10.1093/brain/awn320.
DeKosky ST, Scheff SW: Synapse loss in frontal cortex biopsies in Alzheimer's disease: correlation with cognitive severity. Ann Neurol. 1990, 27: 457-464. 10.1002/ana.410270502.
Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R: Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991, 30: 572-580. 10.1002/ana.410300410.
Ingelsson M, Fukumoto H, Newell KL, Growdon JH, Hedley-Whyte ET, Frosch MP, Albert MS, Hyman BT, Irizarry MC: Early Abeta accumulation and progressive synaptic loss, gliosis, and tangle formation in AD brain. Neurology. 2004, 62: 925-931.
Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ: Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010, 9: 119-128. 10.1016/S1474-4422(09)70299-6.
Savva GM, Wharton SB, Ince PG, Forster G, Matthews FE, Brayne C, Medical Research Council Cognitive Function and Ageing Study: Age, neuropathology, and dementia. N Engl J Med. 2009, 360: 2302-2309. 10.1056/NEJMoa0806142.
Fox NC, Scahill RI, Crum WR, Rossor MN: Correlation between rates of brain atrophy and cognitive decline in AD. Neurology. 1999, 52: 1687-1689.
Frisoni GB, Fox NC, Jack CR, Scheltens P, Thompson PM: The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010, 6: 67-77. 10.1038/nrneurol.2009.215.
Gómez-Isla T, Hollister R, West H, Mui S, Growdon JH, Petersen RC, Parisi JE, Hyman BT: Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer's disease. Ann Neurol. 1997, 41: 17-24. 10.1002/ana.410410106.
Gosche KM, Mortimer JA, Smith CD, Markesbery WR, Snowdon DA: Hippocampal volume as an index of Alzheimer neuropathology: findings from the Nun Study. Neurology. 2002, 58: 1476-1482.
Jack CR, Dickson DW, Parisi JE, Xu YC, Cha RH, O'Brien PC, Edland SD, Smith GE, Boeve BF, Tangalos EG, Kokmen E, Petersen RC: Antemortem MRI findings correlate with hippocampal neuropathology in typical aging and dementia. Neurology. 2002, 58: 750-757.
Silbert LC, Quinn JF, Moore MM, Corbridge E, Ball MJ, Murdoch G, Sexton G, Kaye JA: Changes in premorbid brain volume predict Alzheimer's disease pathology. Neurology. 2003, 61: 487-492.
Braak H, Braak E: Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991, 82: 239-259. 10.1007/BF00308809.
Scheltens P, Leys D, Barkhof F, Huglo D, Weinstein HC, Vermersch P, Kuiper M, Steinling M, Wolters EC, Valk J: Atrophy of medial temporal lobes on MRI in 'probable' Alzheimer's disease and normal ageing: diagnostic value and neuropsychological correlates. J Neurol Neurosurg Psychiatry. 1992, 55: 967-972. 10.1136/jnnp.55.10.967.
Duara R, Loewenstein DA, Potter E, Appel J, Greig MT, Urs R, Shen Q, Raj A, Small B, Barker W, Schofield E, Wu Y, Potter H: Medial temporal lobe atrophy on MRI scans and the diagnosis of Alzheimer disease. Neurology. 2008, 71: 1986-1992. 10.1212/01.wnl.0000336925.79704.9f.
Jack CR, Petersen RC, O'Brien PC, Tangalos EG: MR-based hippocampal volumetry in the diagnosis of Alzheimer's disease. Neurology. 1992, 42: 183-188.
Ashburner J, Friston KJ: Voxel-based morphometry--the methods. Neuroimage. 2000, 11: 805-821. 10.1006/nimg.2000.0582.
Fan Y, Shen D, Davatzikos C: Classification of structural images via high-dimensional image warping, robust feature extraction, and SVM. Med Image Comput Comput Assist Interv. 2005, 8: 1-8. full_text.
Alexander GE, Moeller JR: Application of the scaled subprofile model to functional imaging in neuropsychiatric disorders: a principal component approach to modeling regional patterns of brain function in disease. Human Brain Mapping. 1994, 2: 79-94. 10.1002/hbm.460020108.
Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR: Alzheimer's disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage. 2008, 39: 1186-1197. 10.1016/j.neuroimage.2007.09.073.
Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RS: Automatic classification of MR scans in Alzheimer's disease. Brain. 2008, 131: 681-689. 10.1093/brain/awm319.
Freeborough PA, Fox NC: The boundary shift integral: an accurate and robust measure of cerebral volume changes from registered repeat MRI. IEEE Trans Med Imaging. 1997, 16: 623-629. 10.1109/42.640753.
Jack CR, Shiung MM, Gunter JL, O'Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC: Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004, 62: 591-600.
Thompson PM, Apostolova LG: Computational anatomical methods as applied to ageing and dementia. Br J Radiol. 2007, 80 (Spec No 2): S78-91. 10.1259/BJR/20005470.
Scahill RI, Schott JM, Stevens JM, Rossor MN, Fox NC: Mapping the evolution of regional atrophy in Alzheimer's disease: unbiased analysis of fluid-registered serial MRI. Proc Natl Acad Sci USA. 2002, 99: 4703-4707. 10.1073/pnas.052587399.
Shi F, Liu B, Zhou Y, Yu C, Jiang T: Hippocampal volume and asymmetry in mild cognitive impairment and Alzheimer's disease: meta-analyses of MRI studies. Hippocampus. 2009, 19: 1055-1064. 10.1002/hipo.20573.
Dubois B, Feldman HH, Jacova C, Dekosky ST, Barberger-Gateau P, Cummings J, Delacourte A, Galasko D, Gauthier S, Jicha G, Meguro K, O'Brien J, Pasquier F, Robert P, Rossor M, Salloway S, Stern Y, Visser PJ, Scheltens P: Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007, 6: 734-746. 10.1016/S1474-4422(07)70178-3.
Petersen RC: Mild cognitive impairment. Continuum Lifelong Learning Neurol. 2007, 13: 15-38.
Yuan Y, Gu ZX, Wei WS: Fluorodeoxyglucose-positron-emission tomography, single-photon emission tomography, and structural MR imaging for prediction of rapid conversion to Alzheimer disease in patients with mild cognitive impairment: a meta-analysis. AJNR Am J Neuroradiol. 2009, 30: 404-410. 10.3174/ajnr.A1357.
Fox NC, Freeborough PA: Brain atrophy progression measured from registered serial MRI: validation and application to Alzheimer's disease. J Magn Reson Imaging. 1997, 7: 1069-1075. 10.1002/jmri.1880070620.
Jack CR, Slomkowski M, Gracon S, Hoover TM, Felmlee JP, Stewart K, Xu Y, Shiung M, O'Brien PC, Cha R, Knopman D, Petersen RC: MRI as a biomarker of disease progression in a therapeutic trial of milameline for AD. Neurology. 2003, 60: 253-260. 10.1001/archneur.60.2.253.
Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN: Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: power calculations and estimates of sample size to detect treatment effects. Arch Neurol. 2000, 57: 339-344. 10.1001/archneur.57.3.339.
Vemuri P, Wiste HJ, Weigand SD, Knopman DS, Trojanowski JQ, Shaw LM, Bernstein MA, Aisen PS, Weiner M, Petersen RC, Jack CR, Alzheimer's Disease Neuroimaging Initiative: Serial MRI and CSF Biomarkers in Normal Aging, MCI and AD. Neurology. 2010, 75: 143-151. 10.1212/WNL.0b013e3181e7ca82.
Hashimoto M, Kazui H, Matsumoto K, Nakano Y, Yasuda M, Mori E: Does donepezil treatment slow the progression of hippocampal atrophy in patients with Alzheimer's disease?. Am J Psychiatry. 2005, 162: 676-682. 10.1176/appi.ajp.162.4.676.
Krishnan KR, Charles HC, Doraiswamy PM, Mintzer J, Weisler R, Yu X, Perdomo C, Ieni JR, Rogers S: Randomized, placebo-controlled trial of the effects of donepezil on neuronal markers and hippocampal volumes in Alzheimer's disease. Am J Psychiatry. 2003, 160: 2003-2011. 10.1176/appi.ajp.160.11.2003.
Fox NC, Black RS, Gilman S, Rossor MN, Griffith SG, Jenkins L, Koller M, AN1792(QS-21)-201 Study: Effects of Abeta immunization (AN1792) on MRI measures of cerebral volume in Alzheimer disease. Neurology. 2005, 64: 1563-1572. 10.1212/01.WNL.0000159743.08996.99.
McKeith IG, Dickson DW, Lowe J, Emre M, O'Brien JT, Feldman H, Cummings J, Duda JE, Lippa C, Perry EK, Aarsland D, Arai H, Ballard CG, Boeve B, Burn DJ, Costa D, Del Ser T, Dubois B, Galasko D, Gauthier S, Goetz CG, Gomez-Tortosa E, Halliday G, Hansen LA, Hardy J, Iwatsubo T, Kalaria RN, Kaufer D, Kenny RA, Korczyn A: Diagnosis and management of dementia with Lewy bodies: third report of the DLB Consortium. Neurology. 2005, 65: 1863-1872. 10.1212/01.wnl.0000187889.17253.b1.
Barber R, Gholkar A, Scheltens P, Ballard C, McKeith IG, O'Brien JT: Medial temporal lobe atrophy on MRI in dementia with Lewy bodies. Neurology. 1999, 52: 1153-1158.
Duara R, Barker W, Luis CA: Frontotemporal dementia and Alzheimer's disease: differential diagnosis. Dement Geriatr Cogn Disord. 1999, 10 (Suppl 1): 37-42. 10.1159/000051210.
Galton CJ, Patterson K, Graham K, Lambon-Ralph MA, Williams G, Antoun N, Sahakian BJ, Hodges JR: Differing patterns of temporal atrophy in Alzheimer's disease and semantic dementia. Neurology. 2001, 57: 216-225.
de Leon MJ, DeSanti S, Zinkowski R, Mehta PD, Pratico D, Segal S, Rusinek H, Li J, Tsui W, Saint Louis LA, Clark CM, Tarshish C, Li Y, Lair L, Javier E, Rich K, Lesbre P, Mosconi L, Reisberg B, Sadowski M, DeBernadis JF, Kerkman DJ, Hampel H, Wahlund LO, Davies P: Longitudinal CSF and MRI biomarkers improve the diagnosis of mild cognitive impairment. Neurobiol Aging. 2006, 27: 394-401. 10.1016/j.neurobiolaging.2005.07.003.
Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR, Alzheimer's Disease Neuroimaging Initiative: MRI and CSF biomarkers in normal, MCI, and AD subjects: diagnostic discrimination and cognitive correlations. Neurology. 2009, 73: 287-293. 10.1212/WNL.0b013e3181af79e5.
Walhovd KB, Fjell AM, Brewer J, McEvoy LK, Fennema-Notestine C, Hagler DJ, Jennings RG, Karow D, Dale AM, Alzheimer's Disease Neuroimaging Initiative: Combining MR imaging, positron-emission tomography, and CSF biomarkers in the diagnosis and prognosis of Alzheimer disease. AJNR Am J Neuroradiol. 2010, 31: 347-354. 10.3174/ajnr.A1809.
Brys M, Glodzik L, Mosconi L, Switalski R, De Santi S, Pirraglia E, Rich K, Kim BC, Mehta P, Zinkowski R, Pratico D, Wallin A, Zetterberg H, Tsui WH, Rusinek H, Blennow K, de Leon MJ: Magnetic resonance imaging improves cerebrospinal fluid biomarkers in the early detection of Alzheimer's disease. J Alzheimers Dis. 2009, 16: 351-362.
de Leon MJ, DeSanti S, Zinkowski R, Mehta PD, Pratico D, Segal S, Clark C, Kerkman D, DeBernardis J, Li J, Lair L, Reisberg B, Tsui W, Rusinek H: MRI and CSF studies in the early diagnosis of Alzheimer's disease. J Intern Med. 2004, 256: 205-223. 10.1111/j.1365-2796.2004.01381.x.
Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR, Alzheimer's Disease Neuroimaging Initiative: MRI and CSF biomarkers in normal, MCI, and AD subjects: predicting future clinical change. Neurology. 2009, 73: 294-301. 10.1212/WNL.0b013e3181af79fb.
Wahlund LO, Blennow K: Cerebrospinal fluid biomarkers for disease stage and intensity in cognitively impaired patients. Neurosci Lett. 2003, 339: 99-102. 10.1016/S0304-3940(02)01483-0.
Henneman WJ, Vrenken H, Barnes J, Sluimer IC, Verwey NA, Blankenstein MA, Klein M, Fox NC, Scheltens P, Barkhof F, van der Flier WM: Baseline CSF p-tau levels independently predict progression of hippocampal atrophy in Alzheimer disease. Neurology. 2009, 73: 935-940. 10.1212/WNL.0b013e3181b879ac.
Thomann PA, Kaiser E, Schönknecht P, Pantel J, Essig M, Schröder J: Association of total tau and phosphorylated tau 181 protein levels in cerebrospinal fluid with cerebral atrophy in mild cognitive impairment and Alzheimer disease. J Psychiatry Neurosci. 2009, 34: 136-142.
Herukka SK, Pennanen C, Soininen H, Pirttilä T: CSF Abeta42, tau and phosphorylated tau correlate with medial temporal lobe atrophy. J Alzheimers Dis. 2008, 14: 51-57.
Sluimer JD, Bouwman FH, Vrenken H, Blankenstein MA, Barkhof F, van der Flier WM, Scheltens P: Whole-brain atrophy rate and CSF biomarker levels in MCI and AD: a longitudinal study. Neurobiol Aging. 2008, 31: 758-764. 10.1016/j.neurobiolaging.2008.06.016.
Chou YY, Leporé N, Avedissian C, Madsen SK, Parikshak N, Hua X, Shaw LM, Trojanowski JQ, Weiner MW, Toga AW, Thompson PM, Alzheimer's Disease Neuroimaging Initiative: Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer's disease, mild cognitive impairment and elderly controls. Neuroimage. 2009, 46: 394-410. 10.1016/j.neuroimage.2009.02.015.
Fagan AM, Head D, Shah AR, Marcus D, Mintun M, Morris JC, Holtzman DM: Decreased cerebrospinal fluid Abeta(42) correlates with brain atrophy in cognitively normal elderly. Ann Neurol. 2009, 65: 176-183. 10.1002/ana.21559.
Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Brewer JB, Dale AM, Alzheimer's Disease Neuroimaging Initiative: CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer's disease. J Neurosci. 2010, 30: 2088-2101. 10.1523/JNEUROSCI.3785-09.2010.
Schoonenboom NS, van der Flier WM, Blankenstein MA, Bouwman FH, Van Kamp GJ, Barkhof F, Scheltens P: CSF and MRI markers independently contribute to the diagnosis of Alzheimer's disease. Neurobiol Aging. 2008, 29: 669-675. 10.1016/j.neurobiolaging.2006.11.018.
Schönknecht P, Pantel J, Hartmann T, Werle E, Volkmann M, Essig M, Amann M, Zanabili N, Bardenheuer H, Hunt A, Schröder J: Cerebrospinal fluid tau levels in Alzheimer's disease are elevated when compared with vascular dementia but do not correlate with measures of cerebral atrophy. Psychiatry Res. 2003, 120: 231-238. 10.1016/S0165-1781(03)00197-5.
Mosconi L, Sorbi S, de Leon MJ, Li Y, Nacmias B, Myoung PS, Tsui W, Ginestroni A, Bessi V, Fayyazz M, Caffarra P, Pupi A: Hypometabolism exceeds atrophy in presymptomatic early-onset familial Alzheimer's disease. J Nucl Med. 2006, 47: 1778-1786.
De Santi S, de Leon MJ, Rusinek H, Convit A, Tarshish CY, Roche A, Tsui WH, Kandil E, Boppana M, Daisley K, Wang GJ, Schlyer D, Fowler J: Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiol Aging. 2001, 22: 529-539. 10.1016/S0197-4580(01)00230-5.
Kawachi T, Ishii K, Sakamoto S, Sasaki M, Mori T, Yamashita F, Matsuda H, Mori E: Comparison of the diagnostic performance of FDG-PET and VBM-MRI in very mild Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2006, 33: 801-809. 10.1007/s00259-005-0050-x.
Matsunari I, Samuraki M, Chen WP, Yanase D, Takeda N, Ono K, Yoshita M, Matsuda H, Yamada M, Kinuya S: Comparison of 18F-FDG PET and optimized voxel-based morphometry for detection of Alzheimer's disease: aging effect on diagnostic performance. J Nucl Med. 2007, 48: 1961-1970. 10.2967/jnumed.107.042820.
Ishii K, Soma T, Kono AK, Sofue K, Miyamoto N, Yoshikawa T, Mori E, Murase K: Comparison of regional brain volume and glucose metabolism between patients with mild dementia with lewy bodies and those with mild Alzheimer's disease. J Nucl Med. 2007, 48: 704-711. 10.2967/jnumed.106.035691.
Hinrichs C, Singh V, Mukherjee L, Xu G, Chung MK, Johnson SC, Alzheimer's Disease Neuroimaging Initiative: Spatially augmented LPboosting for AD classification with evaluations on the ADNI dataset. Neuroimage. 2009, 48: 138-149. 10.1016/j.neuroimage.2009.05.056.
Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergström M, Savitcheva I, Huang GF, Estrada S, Ausén B, Debnath ML, Barletta J, Price JC, Sandell J, Lopresti BJ, Wall A, Koivisto P, Antoni G, Mathis CA, Långström B: Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004, 55: 306-319. 10.1002/ana.20009.
Driscoll I, Zhou Y, An Y, Sojkova J, Davatzikos C, Kraut MA, Ye W, Ferrucci L, Mathis CA, Klunk WE, Wong DF, Resnick SM: Lack of association between (11)C-PiB and longitudinal brain atrophy in non-demented older individuals. Neurobiol Aging. 2010,
Storandt M, Mintun MA, Head D, Morris JC: Cognitive decline and brain volume loss as signatures of cerebral amyloid-beta peptide deposition identified with Pittsburgh compound B: cognitive decline associated with Abeta deposition. Arch Neurol. 2009, 66: 1476-1481. 10.1001/archneurol.2009.272.
Chételat G, Villemagne VL, Bourgeat P, Pike KE, Jones G, Ames D, Ellis KA, Szoeke C, Martins RN, O'Keefe GJ, Salvado O, Masters CL, Rowe CC, Australian Imaging Biomarkers and Lifestyle Research Group: Relationship between atrophy and beta-amyloid deposition in Alzheimer disease. Ann Neurol. 2010, 67: 317-324.
Bourgeat P, Chételat G, Villemagne VL, Fripp J, Raniga P, Pike K, Acosta O, Szoeke C, Ourselin S, Ames D, Ellis KA, Martins RN, Masters CL, Rowe CC, Salvado O, AIBL Research Group: Beta-amyloid burden in the temporal neocortex is related to hippocampal atrophy in elderly subjects without dementia. Neurology. 2010, 74: 121-127. 10.1212/WNL.0b013e3181c918b5.
Jack CR, Lowe VJ, Senjem ML, Weigand SD, Kemp BJ, Shiung MM, Knopman DS, Boeve BF, Klunk WE, Mathis CA, Petersen RC: 11C PiB and structural MRI provide complementary information in imaging of Alzheimer's disease and amnestic mild cognitive impairment. Brain. 2008, 131: 665-680. 10.1093/brain/awm336.
Archer HA, Edison P, Brooks DJ, Barnes J, Frost C, Yeatman T, Fox NC, Rossor MN: Amyloid load and cerebral atrophy in Alzheimer's disease: an 11C-PIB positron emission tomography study. Ann Neurol. 2006, 60: 145-147. 10.1002/ana.20889.
Scheinin NM, Aalto S, Koikkalainen J, Lötjönen J, Karrasch M, Kemppainen N, Viitanen M, Någren K, Helin S, Scheinin M, Rinne JO: Follow-up of [11C]PIB uptake and brain volume in patients with Alzheimer disease and controls. Neurology. 2009, 73: 1186-1192. 10.1212/WNL.0b013e3181bacf1b.
Teipel SJ, Ewers M, Wolf S, Jessen F, Kölsch H, Arlt S, Luckhaus C, Schönknecht P, Schmidtke K, Heuser I, Frölich L, Ende G, Pantel J, Wiltfang J, Rakebrandt F, Peters O, Born C, Kornhuber J, Hampel H: Multicentre variability of MRI-based medial temporal lobe volumetry in Alzheimer's disease. Psychiatry Res. 2010, 182: 244-250. 10.1016/j.pscychresns.2010.03.003.
Hampel H, Bürger K, Pruessner JC, Zinkowski R, DeBernardis J, Kerkman D, Leinsinger G, Evans AC, Davies P, Möller HJ, Teipel SJ: Correlation of cerebrospinal fluid levels of tau protein phosphorylated at threonine 231 with rates of hippocampal atrophy in Alzheimer disease. Arch Neurol. 2005, 62: 770-773. 10.1001/archneur.62.5.770.
Schoonenboom SN, Visser PJ, Mulder C, Lindeboom J, Van Elk EJ, Van Kamp GJ, Scheltens PH: Biomarker profiles and their relation to clinical variables in mild cognitive impairment. Neurocase. 2005, 11: 8-13. 10.1080/13554790490896785.
Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, Jack CR, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Borowski B, Shaw LM, Trojanowski JQ, Fleisher AS, Harvey D, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM, Alzheimer's Disease Neuroimaging Initiative: Alzheimer's disease neuroimaging initiative: a one-year follow up study using tensor-based morphometry correlating degenerative rates, biomarkers and cognition. Neuroimage. 2009, 45: 645-655. 10.1016/j.neuroimage.2009.01.004.
Schuff N, Woerner N, Boreta L, Kornfi eld T, Shaw LM, Trojanowski JQ, Thompson PM, Jack CR, Weiner MW, Alzheimer's Disease Neuroimaging Initiative: MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain. 2009, 132: 1067-1077. 10.1093/brain/awp007.
Fjell AM, Walhovd KB, Fennema-Notestine C, McEvoy LK, Hagler DJ, Holland D, Blennow K, Brewer JB, Dale AM, the Alzheimer's Disease Neuroimaging Initiative: Brain atrophy in healthy aging is related to CSF levels of A{beta}1-42. Cereb Cortex. 2010, 20: 2069-2079. 10.1093/cercor/bhp279.
Yamaguchi S, Meguro K, Itoh M, Hayasaka C, Shimada M, Yamazaki H, Yamadori A: Decreased cortical glucose metabolism correlates with hippocampal atrophy in Alzheimer's disease as shown by MRI and PET. J Neurol Neurosurg Psychiatry. 1997, 62: 596-600. 10.1136/jnnp.62.6.596.
Ishii K, Sasaki H, Kono AK, Miyamoto N, Fukuda T, Mori E: Comparison of gray matter and metabolic reduction in mild Alzheimer's disease using FDG-PET and voxel-based morphometric MR studies. Eur J Nucl Med Mol Imaging. 2005, 32: 959-963. 10.1007/s00259-004-1740-5.
Samuraki M, Matsunari I, Chen WP, Yajima K, Yanase D, Fujikawa A, Takeda N, Nishimura S, Matsuda H, Yamada M: Partial volume effect-corrected FDG PET and grey matter volume loss in patients with mild Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2007, 34: 1658-1669. 10.1007/s00259-007-0454-x.
Chételat G, Desgranges B, Landeau B, Mézenge F, Poline JB, de la Sayette V, Viader F, Eustache F, Baron JC: Direct voxel-based comparison between grey matter hypometabolism and atrophy in Alzheimer's disease. Brain. 2008, 131: 60-71. 10.1093/brain/awm288.
Walhovd KB, Fjell AM, Amlien I, Grambaite R, Stenset V, Bjørnerud A, Reinvang I, Gjerstad L, Cappelen T, Due-Tønnessen P, Fladby T: Multimodal imaging in mild cognitive impairment: metabolism, morphometry and diffusion of the temporal-parietal memory network. Neuroimage. 2009, 45: 215-223. 10.1016/j.neuroimage.2008.10.053.
Morbelli S, Piccardo A, Villavecchia G, Dessi B, Brugnolo A, Piccini A, Caroli A, Frisoni G, Rodriguez G, Nobili F: Mapping brain morphological and functional conversion patterns in amnestic MCI: a voxel-based MRI and FDG-PET study. Eur J Nucl Med Mol Imaging. 2010, 37: 36-45. 10.1007/s00259-009-1218-6.
Acknowledgements
PV receives support from the Robert H. Smith Family Foundation Research Fellowship. CRJ receives support from the National Institute on Aging - AG11378 (PI), P50-AG16574 (Co-I), and U01 AG024904-01 (Co-I) - and the Alexander Family Alzheimer's Disease Research Professorship of the Mayo Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
CRJ serves as a consultant for Eli Lilly and Company (Indianapolis, IN, USA) and Elan Corporation (Dublin, Ireland) and is an investigator in clinical trials sponsored by Baxter (Deerfield, IL, USA) or Pfizer Inc. (New York, NY, USA) and holds stock in GE Healthcare (Waukesha, WI, USA). PV declares that she has no competing interests.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
About this article
Cite this article
Vemuri, P., Jack, C.R. Role of structural MRI in Alzheimer's disease. Alz Res Therapy 2, 23 (2010). https://doi.org/10.1186/alzrt47
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/alzrt47