Figure 2. Principal component analysis of two populations.
(A) Consider a sample of 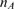 individuals from population A (indicated by the red circle) and
individuals from population A (indicated by the red circle) and  from population B (indicated by the blue circle), where the two populations have the same effective population size of
from population B (indicated by the blue circle), where the two populations have the same effective population size of  and are both derived from a single ancestral population, also of size
and are both derived from a single ancestral population, also of size  , with the split happening a time
, with the split happening a time  in the past. (B) The expected locations of these two sets of samples on the first PC is defined by the time since divergence (the Euclidean distance between the samples is
in the past. (B) The expected locations of these two sets of samples on the first PC is defined by the time since divergence (the Euclidean distance between the samples is 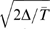 ) (see text for definitions) and the relative sample size from the populations, with the larger sample lying closer to the origin. Defining
) (see text for definitions) and the relative sample size from the populations, with the larger sample lying closer to the origin. Defining  , the relative location of the two populations on the first PC are
, the relative location of the two populations on the first PC are  for samples from population A and
for samples from population A and  for samples from population B (note that the sign is arbitrary). (C) To investigate the effect of finite genome size simulations were carried out for the model shown in part A with 80 genomes sampled from population A, 20 from population B and a split time of 0.02
for samples from population B (note that the sign is arbitrary). (C) To investigate the effect of finite genome size simulations were carried out for the model shown in part A with 80 genomes sampled from population A, 20 from population B and a split time of 0.02  generations (
generations ( ) and between
) and between  and
and  SNPs. Lines indicate the analytical expectation. A jitter has been added to the x-axis for clarity. Note that the separation of samples with 10 SNPs does not correlate with population and simply reflects random clustering arising from the small numbers of SNPs.
SNPs. Lines indicate the analytical expectation. A jitter has been added to the x-axis for clarity. Note that the separation of samples with 10 SNPs does not correlate with population and simply reflects random clustering arising from the small numbers of SNPs.

