Abstract
Small RNAs (sRNAs) are hypothesized to contribute to hybrid vigor because they maintain genome integrity, contribute to genetic diversity, and control gene expression. We used Illumina sequencing to assess how sRNA populations vary between two maize inbred lines (B73 and Mo17) and their hybrid. We sampled sRNAs from the seedling shoot apex and the developing ear, two rapidly growing tissues that program the greater growth of maize hybrids. We found that parental differences in siRNAs primarily originate from repeat regions. Although the maize genome contains greater number and complexity of repeats compared with Arabidopsis or rice, we confirmed that, like these simpler plant genomes, 24-nt siRNAs whose abundance differs between maize parents also show a trend of down-regulation following hybridization. Surprisingly, hybrid vigor is fully maintained when 24-nt siRNAs are globally reduced by mutation of the RNA-dependent RNA polymerase 2 encoded by modifier of paramutation1 (mop1). We also discovered that 21–22-nt siRNAs derived from a number of distinct retrotransposon families differentially accumulate between B73 and Mo17 as well as their hybrid. Thus, maize possesses a unique source of genetic variation for regulating transposons and genes at a genomic scale, which may contribute to its high degree of observed heterosis.
Keywords: transposon regulation, 22-nucleotide RNAs
In animals, plants, and fungi, hybridization frequently produces offspring more vigorous than their parents. The phenomenon of hybrid vigor, or heterosis, depends on genetic variation between parents and altered genetic states in their offspring. Shull, who conducted the first genetic analyses of the phenomenon in maize (1), stressed 60 y ago that multiple mechanisms likely contribute to different examples of hybrid vigor (2). Advancements in our understanding of genome structure (3, 4) and gene regulation make this statement even more relevant today. New sources of genetic variation continue to be uncovered (5), along with regulatory systems whereby hybridization combines variants with nonadditive phenotypic effects on growth, metabolism, and environmental response.
Small RNAs (sRNAs) regulate gene expression and maintain genome integrity (6, 7), both of which are impacted by hybridization. The relative ease of self-propagation, hybridization, and developmental staging of different generations in plants makes it feasible to investigate the variation in sRNAs between parents and their progeny. Studies in Arabidopsis (8–10), rice (11), and wheat (12) have used sRNA sequencing because the technology catalogues the various size classes of sRNAs and the relative abundance (i.e., normalized reads of a sequence signature) of specific sRNAs or sRNA classes among genotypes. Collectively, these studies find genetic variation in 24-nt siRNAs, reductions of 24-nt siRNAs following hybridization, and nonadditive expression of key regulatory microRNAs (miRNAs) and their gene targets. Each of these observed properties for sRNAs fit the genetic principles known and hypothesized to contribute to hybrid vigor in plants (13).
The magnitude of hybrid vigor in maize is relatively high and observed throughout its life cycle, which has made the species an excellent model for studying the phenomenon (14). Moreover, hybrid maize has been an important application of heterosis. The maize genome differs from model plant species such as rice and Arabidopsis by the presence of many classes of high-copy repeats, particularly transposable elements (TEs), whose activities are silenced by siRNAs. Accordingly, maize possesses unique features of sRNA production, such as an abundant class of 22-nt siRNAs that is derived from retrotransposons through a pathway distinct from the generation of 24-nt siRNAs by RNA-dependent RNA polymerase 2 (RDR2) (15). To learn more about how hybridization impacts sRNAs in maize, we sequenced sRNAs from the seedling shoot apex and developing ear of two maize inbred lines (B73 and Mo17) and their hybrids. We chose these genotypes because of (i) their high degree of hybrid vigor, (ii) they represent the major heterotic groups deployed in historical and current North American corn breeding, and (iii) the extensive genomics resources that exist for the parental lines. We also investigated whether the genetic contribution of RDR2-mediated amplification of transcriptional gene silencing contributes to the hybrid vigor displayed by B73×Mo17.
Results
sRNA Sequencing of B73, Mo17, and Their Hybrids.
To investigate differences in sRNA profiles between maize parents and their hybrids, we sampled (i) seedling shoot apex tissues at 11 d after sowing (DAS) and (ii) developing ear tissues at the time when the 12th leaf had fully expanded (i.e., V12), from B73, Mo17, and their hybrids (Fig. 1 A and B). The hybrids showed heterosis for size when the tissues were sampled (Fig. S1). We chose to study the shoot apex because it is enriched for meristematic tissue, where cell proliferation occurs, rates of organ initiation are determined, and organ size is specified. We also examined the developing ear because it is also enriched in meristematic tissue and is undergoing rapid growth, and also because the mature ear shows the highest degree of heterosis (16). Furthermore, comparison of sRNA profiles for developing shoots and ears may reveal if sRNA production or activity is influenced by accumulated physiological differences that occur during vegetative development.
Fig. 1.
Summary of actively growing and highly proliferative tissues investigated for maize inbred and hybrid sRNA sequencing experiment. (A) At 11 DAS, tissues enriched for the shoot apex were collected by removing emerged leaves and sampling the bottom 1 cm of the remaining leaf tissue. (B) When the shoot had elongated 12 fully-expanded leaves (i.e., at V12), the top developing ear was excised.
sRNA libraries were made for each genotype from RNA extracted from pooled shoot apices or developing ears and sequenced by using the Illumina sequencing by synthesis platform (Materials and Methods) (Table S1). After processing the sRNA data and combining sequences across the libraries (Matrials and Methods), we identified a set of 95,665 distinct sRNAs representing 3,134,719 reads for the shoot apex and a set of 118,625 distinct sRNAs representing 3,132,802 reads for the developing ear (Dataset S1). As expected, more sRNAs catalogued from a given parent matched its own genome sequences compared with those from the other parent (Table S1).
The sRNA populations are enriched for repeat-associated siRNAs (rasiRNAs) as shown by the large percentages of retrotransposon derived siRNAs, sRNAs matching ribosomal DNA elements, and siRNAs with a high copy number (>10 locations) in the B73 genome (Fig. S2A). miRNAs account for 16% of the shoot apex sRNA population but only 2% of the developing ear sRNA population (Fig. S2A). The miRNA profiles of the two tissues differ dramatically and consist of a few highly abundant miRNAs (Fig. S2B). Some miRNAs appear to accumulate nonadditively, most notably microRNA168 (miR168), which increased approximately twofold in the hybrids for both tissues. However, in additional replicated experiments using a quantitative real-time PCR (qRT-PCR) assay for miR168, we found that it accumulates to similar levels in the parents and hybrid during the course of early shoot and ear development (Fig. S3A).
The expression of microRNA156 (miR156) declines during early maize shoot development to promote vegetative phase change (17). Mo17 and B73×Mo17 both transition to the adult phase earlier than B73 (Fig. S3B). Thus, we were initially surprised to see that miR156 is more abundant in Mo17 compared with B73 at 11 DAS, whereas both reciprocal hybrids had lower abundance than B73. After controlling for differences in plastochron length (i.e., the rate of leaf initiation), the expression of miR156 at the three-leaf stage did in fact reflect the shorter juvenile phase of Mo17 and the B73×Mo17 hybrid relative to B73 (Fig. S3C). The differences among the genotypes disappeared as miR156 levels declined and shoots transitioned to the adult vegetative phase. The data demonstrate that observed differences in the rate of development for hybrids compared with parents can be detected as differences in expression of miRNAs that control the rate of organ initiation and shoot maturation.
Hybrids Combine Parental Differences in siRNA Populations.
We found that differences between parents and hybrids in sRNA populations primarily result from the hybrids inheriting distinct siRNAs from each parent. The sRNA length profile is similar to those previously reported in maize (15, 18) and does not differ among genotypes (Fig. S2C), which indicates that hybridization does not alter biogenesis of different sRNA size classes. The relative difference in abundance between 24-nt sRNAs and the other lengths is larger in the ear than in the shoot apex, possibly because the expression of mop1 is higher in the ear compared with the seedling (18).
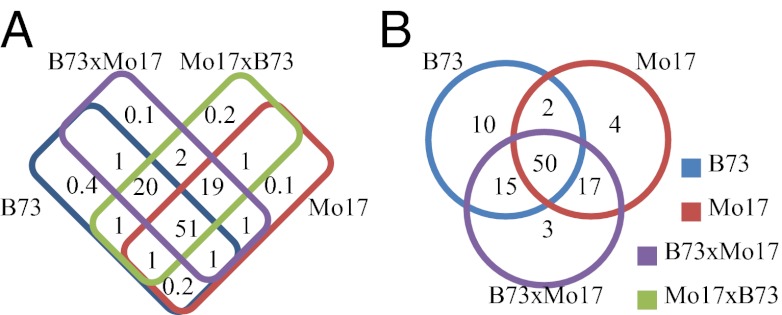
To investigate siRNAs, we removed sRNAs matching miRNAs, ribosomal DNA, or tRNA from the datasets and retained only those that mapped to either the B73 or Mo17 genome and had the characteristic siRNA length (21–24-nt). The vast majority (ear, 82%; shoot apex, 90%) of this siRNA population was sampled from each of the genotypes or in one of the parents and hybrids (Fig. 2). Few siRNAs are unique to one parent, present in parents but not hybrids, or found in hybrids but not parents. Therefore, hybridization does not create new siRNAs; instead, hybrids possess a more complex siRNA population than either parent by inheriting siRNAs from both parents.
Fig. 2.
Global differences in sRNAs between parents and hybrids result from parents passing on different populations of distinct siRNAs. Venn diagrams show percentage of total 21- to 24-nt siRNA abundance accounted for by each genotypic group for shoot apex (A) and developing ear (B).
To further characterize the inheritance of parental differences in sRNAs, we calculated d/a ratios [ratio of dominance (hybrid relative to midparent) to additive (midparent) gene action] (19) for 21–24-nt siRNAs that map to only one parent’s genome and were observed from RNA of that genotype at an abundance of at least five reads per million (rpm), but not detected in the other parent. The distributions of d/a values for the shoot apex suggest that these siRNAs are primarily inherited in additive manner; however, in the developing ear, the d/a values are strongly biased to below midparent levels (Fig. S4). Most of the siRNAs exhibiting this behavior are 24-nt (B73, 91%; Mo17, 92%). We also observed this bias in developing ear samples taken from low nitrogen field plots (Fig. S4 E and F). Again, most of the siRNAs exhibiting this behavior are 24-nt (B73, 92%; Mo17, 93%).
Parental Differences in siRNAs Primarily Originate from Repeats.
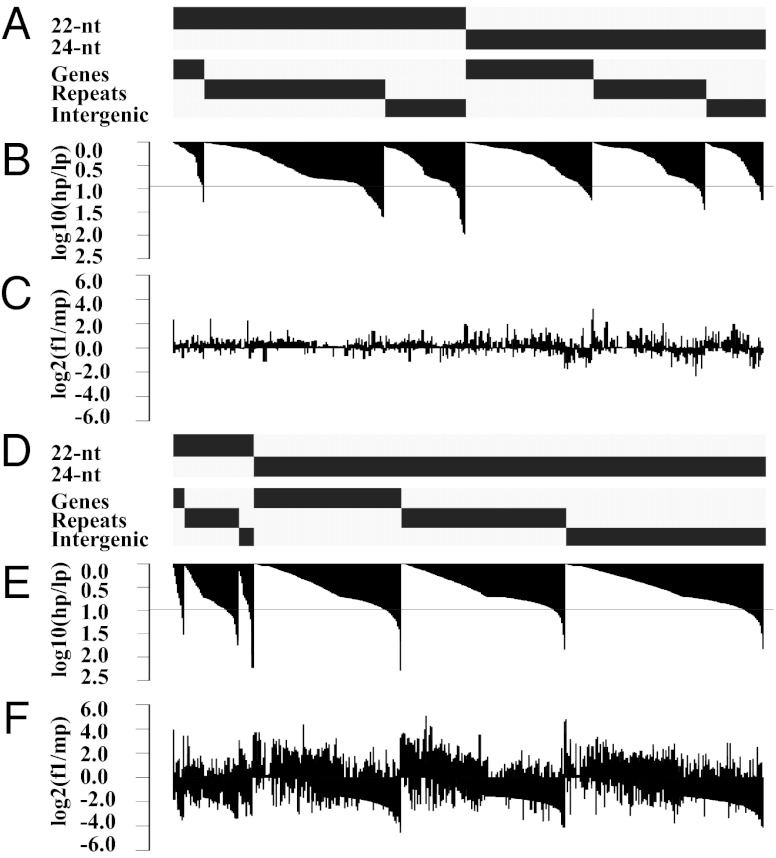
We used an approach similar as Johnson et al. (20) to identify the types of genetic features in which parental differences in siRNAs originate. We grouped 21- to 24-nt siRNAs that overlapped (≤100 bp) and mapped to both the B73 and the Mo17 genomes into clusters based on their location within the B73 genome (Materials and Methods). The abundance of siRNAs that mapped to more than one location was repeat-normalized before the summation of a clusters’ total siRNA abundance (i.e., rpm-repnorm). The siRNA clusters may contain sequences ranging in size from 21 to 24 nt but are referred to by their most common siRNA length (21, 22, or 24-nt) and are characterized by the genetic feature annotated at the location of their match in the B73 genome. For each tissue, we analyzed the siRNA clusters that had an abundance of at least 5 rpm-repnorm in one of the genotypes. This data are available in Dataset S1. The 21-nt clusters accounted for less than 1% of the clusters in both tissues.
In the shoot and ear, the proportion of 21-nt and 22-nt siRNAs in a cluster are positively correlated (shoot, r = 0.51; ear, r = 0.46; P < 0.0001), whereas the proportions of 21-nt and 22-nt siRNAs in a cluster are negatively correlated with 24-nt siRNAs (shoot, r = −0.71 and r = −0.95, respectively; ear, r = −0.68 and r = −0.79, respectively; P < 0.0001). The 22-nt clusters have a longer mean length (668 bp) and are more abundant (7 rpm) than the 24-nt clusters (89 bp and 5 rpm; Wilcoxon rank-sum P < 0.0001). The 24-nt clusters are more likely to be located near genes than 22-nt clusters, which are instead found in repetitive sequences (Fig. 3 A and D). To investigate clusters exhibiting large parental differences, we ordered the clusters by their degree of parental fold change and selected clusters within the top 10% of these values (Fig. 3 B and E). In both tissues, these clusters primarily map to repeats (Fig. 3 A and D). When additional replicate sRNA sequence datasets were examined for the parental genotypes, we detected clusters in the same genomic location, with the same high parent and in the top 10% of the parental differences for 39% and 25% of these shoot apex and developing ear clusters, respectively. Comparing the top 10% clusters for all four sRNA sequencing datasets, we found 18 siRNA clusters in the shoot apex that matched 20 clusters in the developing ear. Based on proximity (<250 bp), we collapsed these clusters into 10 genomic regions. Interestingly, eight of these regions consisted only of 22-nt clusters and are located in genomic intervals containing sequences annotated as high-copy retrotransposon families (Table S2).
Fig. 3.
Parental differences in siRNA regions primarily originate from repeats and deviate less from midparent in the shoot apex compared with the developing ear. Shown are the 22-nt and 24-nt siRNA clusters with at least 5 rpm-repnorm for shoot apex (A–C; n = 1,306) and developing ear (D–F; n = 5,110). Clusters are arranged in ascending order of parental fold change. (A and D) Classification of clusters based on type and genetic feature. (B and E) Degree of parental difference for siRNA clusters (log10 of high parent abundance divided by low parent abundance). Clusters below the horizontal gray line have parental differences that fall within the top 10% of the values for all of the clusters (shoot apex, 8.9-fold; ear, 9.5-fold). (C and F) Deviation from midparent values for siRNA clusters (log2 of F1 abundance divided by midparent abundance).
We calculated the deviation from the midparent abundance for each siRNA cluster to investigate how the clusters behave following hybridization. Fig. 3 C and F show deviation from midparent values for the clusters arranged in increasing order of the parental fold change of the clusters. In the shoot apex, siRNA clusters appear to be inherited in an additive manner (Fig. 3C). In contrast, siRNA clusters in the ear show larger deviations above and below midparent values, but trend to below midparent levels as the degree of difference in siRNA cluster abundance between the parents increases (Fig. 3F). A similar trend was observed for the developing ear samples taken from the low nitrogen plots (Fig. S5C). In both developing ear datasets, this trend is more strongly observed for the 24-nt clusters.
Parental Differences in Retrotransposon siRNA Activity Are Driven by 21-nt and 22-nt siRNAs.
To further investigate global differences in siRNA abundance for retrotransposon families between the parents, we mapped 21-nt, 22-nt, and 24-nt siRNAs that perfectly matched the B73 or Mo17 genomes onto the characterized retrotransposons present in the Zea repeats database. Fig. 4 shows the retrotransposon families that had an abundance of at least 100 rpm in one of the genotypes. The production of specific siRNA lengths and the overall abundance of siRNAs from these retrotransposon families are similar across the two tissues. The ji and cinful families have the highest total abundance. The families can be grouped into those that produce primarily 22-nt siRNAs (cinful, rire1, giepum, ji, misfit), those that produce both 22-nt and 24-nt siRNAs (zeon, grande), and those that produce primarily 24-nt siRNAs (huck1, milt, opie).
Fig. 4.
Parental differences in retrotransposon siRNA activity are driven by 21-nt and 22-nt siRNAs. siRNA profiles are displayed for shoot apex (A) and developing ear (B). The total abundance column displays the relative abundance of 21-, 22-, and 24-nt siRNAs matching maize characterized retrotransposon families (at most 1-bp mismatch). For each family, abundance is partitioned by genotype and siRNA length.
Parental differences in the abundance of siRNAs for cinful, zeon, rire1, giepum, grande, and ji were consistent in both tissues. We used a χ2 test to determine if these differences between B73 and Mo17 were associated with a particular length in siRNA. We used the tissues as biological replicates for the parental genotypes, requiring the significant difference to be observed in the same direction in both tissues. The difference in abundance for cinful, grande, and ji between B73 and Mo17 is contingent on the 21-nt and 22-nt lengths, whereas the parental difference for giepum, rire1, and zeon1 is contingent on only the 22-nt length (Bonferroni-corrected P < 0.01). Significant differences between B73 and Mo17 for retrotransposon families were observed for 24-nt siRNAs, but they were either not significant in both tissues or occurred in different directions in the tissues. In additional sRNA sequencing experiments, we again observed these significant contingencies between the parental difference in abundance and siRNA length (21-nt, cinful, ji; 22-nt, cinful, rire1, giepum, grande, ji; Fig. S6). Therefore, the differences in siRNA abundance for these retrotransposon families primarily result from 21- to 22-nt siRNAs.
The parental differences in abundance for 21- to 22-nt siRNAs from specific retrotransposon families may reflect differential transcription of source sequences and subsequent processing into sRNAs, or could indicate down-regulation of complementary mRNA targets by posttranscriptional activities. We performed qRT-PCR to differentiate between these two scenarios for the cinful family, which had a greater abundance of 21-nt and 22-nt siRNAs in B73. In the shoot apex, we found that cinful mRNA levels increase from the three- to four-leaf stage (Fig. S3D). At the four- and five-leaf stages, B73 has significantly higher levels of cinful mRNA than Mo17, and the hybrid appears to track the higher parent. For the V10 to V13 growth stages in the developing ear, we found that B73 has higher levels of cinful mRNA than Mo17, and the hybrid has levels between the parents. Thus, at least for cinful in B73, 21- to 22-nt siRNAs accumulate in proportion to retrotransposon-derived mRNAs.
Loss of mop1 Does Not Suppress Hybrid Vigor for B73×Mo17.
RDR genes function as the amplification components of RNA silencing pathways, producing dsRNA from single-stranded precursors to sustain silencing (21). Loss of mop1, an RDR2 orthologue in maize (22), has drastic phenotypic effects, such as stunting, delayed flowering, and feminization of tassels (23). The mop1 mutation dramatically reduces the abundance of a large population of 24-nt siRNAs, but does not affect 22-nt siRNAs (15). Therefore, the mutation provides a genetic system to test the contribution of mop1-dependent 24-nt siRNAs for hybrid vigor in maize.
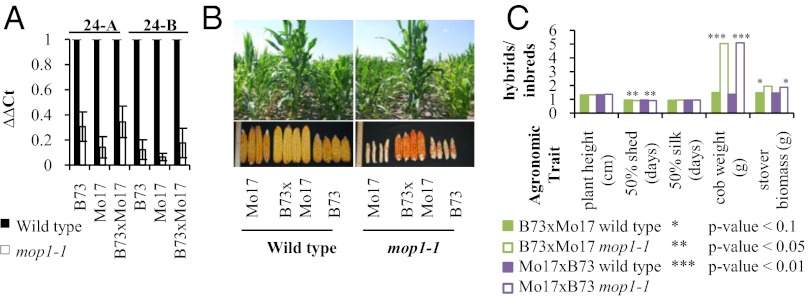
The mop1-1 loss of function allele (22) was introgressed into the B73 and Mo17 inbred backgrounds, and reciprocal hybrids from WT and mop1-1 mutant parents were generated. The parental inbreds and reciprocal hybrids were each grown in a replicated field trial. To verify the expected effects of mop1-1 on 24-nt siRNA accumulation, we used qRT-PCR to measure the amount of two 24-nt siRNAs previously documented to have reduced abundance in the developing ear from mop1-1 mutant plants (15). Fig. 5A shows that the levels of the two 24-nt siRNAs are significantly reduced in the developing ear for the mop1-1 mutants. We found that, for all genotypes, mop1-1 significantly reduces plant height and cob weight, and also delays flowering (Table S3). Stover biomass significantly decreases for the mutant compared with normal inbreds. Although mop1-1 impacted the mean genotypic values of these traits in the parents, the mutation did not suppress the heterotic behavior of B73×Mo17 or Mo17×B73. Vigor for vegetative and reproductive tissues was readily observed for mutant hybrids (Fig. 5B). The hybrid performance observed in the mutant is no less than the WT plants and is even enhanced for days to 50% shed, cob weight, and stover biomass (Fig. 5C).
Fig. 5.
Loss of mop1-1 reduces 24-nt siRNAs in the developing ear but does not suppress hybrid vigor for B73×Mo17. (A) ΔΔCt values for siRNAs 24-A and 24-B using microRNA172 as the reference. RNAs were assayed from developing tissue (top ear from V10–V12 plant growth stage) from WT and mop1-1 field grown plants. Error bars represent ±2 SEM of the ΔΔCt values for four individual ears. (B) Vegetative and reproductive growth for WT and mop1-1 mutant B73×Mo17 hybrids compared with their parents. (C) Hybrid-to-inbred ratio for mean agronomic trait values. For each genotype, the height of 20 plants and cob weight of 25 plants were measured, and five replicate rows were measured for flowering traits and stover biomass.
Discussion
Core Components of sRNA Biogenesis and Hybrid Vigor.
Dosage changes in key regulatory genes have been proposed to explain the nonadditive phenotypes of hybridization (13). sRNAs are good candidates for such factors because they regulate gene expression via binding to complementary RNAs. However, the results obtained from sRNA sequencing indicate that key components to sRNA biogenesis do not change following hybridization of B73 and Mo17, and conversely, dramatic changes in the production of RDR2-dependent 24-nt siRNAs have little impact on the degree of hybrid vigor displayed by B73×Mo17. Among miRNAs, we might expect changes in miR168 abundance to have the greatest molecular effects because it functions as a core regulator of sRNA accumulation through its posttranscriptional regulation of ARGONAUTE 1 (AGO1) (24). In wheat, Kenan-Eichler et al. (12) attributed a global decrease in 24-nt siRNAs following allopolyploidization to a twofold increase in miR168 abundance in the allopolyploid. From our sRNA sequencing data, we also observed an approximate twofold increase in miR168 abundance in the hybrids for both tissues. However, by using a more sensitive qRT-PCR assay and greater biological replication, we found that miR168 does not differentially accumulate among B73, Mo17, and their hybrid (Fig. S3A), which is consistent with our finding that 24-nt siRNAs are not globally decreased relative to other sizes in the hybrids (Fig. S2C). Although there is evidence that suggests miRNAs are nonadditively expressed following hybridization in plants (8, 11), another study did not find strong examples (9). It is important to remember that the increased size of hybrids relative to their parents occurs in the context of normal developmental programs for organ initiation, morphogenesis, and differentiation. This principle led East (25) to suggest that genes controlling development are not important to hybrid vigor. Because many miRNAs regulate developmental processes that continue to operate normally in hybrids, they might not be expected to be key drivers of hybrid vigor. However, as we demonstrate here for miR156, miRNAs can certainly respond to hybridization (Fig. S2B).
In maize, loss of the RDR2 orthologue mop1 reduces global levels of 24-nt siRNAs (15) alters the expression of thousands of genes and TEs (26) and has drastic consequences for the growth and development of inbred lines (Table S3) (23). However, we found that loss of mop1 does not result in a decrease of hybrid vigor displayed by B73×Mo17 or Mo17×B73 for vegetative or reproductive traits (Fig. 5 B and C). The magnitude of hybrid vigor was even enhanced for days to 50% anthesis, cob weight, and stover biomass. Because the effects of mutations frequently depend on genetic background, we caution direct comparisons of our results with findings from other studies of mop1 mutants (15, 26). However, we did confirm via qRT-PCR that 24-nt siRNAs were reduced in each of the genotypes also homozygous for mop1-1 (Fig. 5A). Therefore, we conclude that mop1-dependent 24-nt siRNAs are not required for the hybrid vigor of measured traits for this specific cross.
Hybridization and 24-nt siRNAs.
We found that hybridization does not alter sRNA populations globally in either of two rapidly developing tissues that dictate organ number and size (Fig. S2C). Instead, hybridization combines parental differences in siRNAs, producing an offspring that is more complex than either parent. Our sRNA sequencing results show that the hybrid inherits nearly all the differences in siRNA populations between B73 and Mo17 (Fig. 2). These 21- to 24-nt siRNAs mainly match repeat regions of the B73 genome (Figs. 3 and 4). Most of the 24-nt siRNAs are likely involved in the transcriptional regulation of TEs through RNA-directed DNA methylation (6), but they could also influence gene expression in cis, as 24-nt siRNA regions were found to occur within or near genes (Fig. 3). In the ear but not the shoot apex, we note that the 24-nt siRNAs and siRNA clusters that differ between parents tend to accumulate to levels below midparent levels (Fig. 3 and Figs. S4 and S5). We attribute the greater degree of nonadditive inheritance in the ear to its more heterogeneous population of cells produced later in development, when cumulative physiological effects may have greater impact (13).
Similar to this work, reductions of 24-nt siRNAs following hybridization or polyploidization have been documented in a number of plant species (8, 9, 11, 12). If this is a general trend, a simple explanation for our observation that mop1 mutants do not display reduced hybrid vigor may be that reductions in RDR2 produce a similar regulatory outcome to hybridization, namely reduced production of 24-nt siRNAs. Prior studies have reported global and local reductions in 24-nt siRNAs, which could reflect different approaches to processing and analysis of the raw sRNA sequencing data or the different tissues investigated. In Arabidopsis thaliana, Groszmann et al. (9) connected the reduction in 24-nt siRNAs to changes in gene expression in the hybrid that were mediated through a loss of DNA methylation. If hybridization functions to reduce or reset parental differences in 24-nt siRNAs, presumably, siRNA accumulation and epigenetic regulation are reestablished in some manner in subsequent generations of inbreeding. Recently, de novo variation in 21- to 24-nt siRNA abundance was found in F2-derived lines of a cross between a wild and a modern tomato cultivar and in introgression lines of the wild germplasm into the modern background (27). The authors note that the reestablishment of siRNAs in subsequent generations following hybridization could have important consequences in plant breeding if they have phenotypic effects.
Retrotransposons and Posttranscriptional Regulatory Variation.
Unlike previous studies comparing sRNAs between parents and their hybrids in plants (8–12), we found significant parental variation in 21- to 22-nt siRNAs derived from specific retrotransposon families (Fig. 4, Fig. S6, and Table S2). This difference likely reflects our choice to investigate actively dividing tissues, whereas other studies (8–12) sequenced sRNAs from mature tissues or even whole seedlings, in which transcription of TEs is less likely to be observed. For example, Shen et al. (10) noted at least a fivefold enrichment in sRNA associated with genes compared with those associated with TEs. This difference between the studies may also reflect the different impact that TEs have played on the genome biology of the species. In fact, very few TEs have been identified in Arabidopsis that produce 21- to 22-nt siRNAs (28). As we observed from our data (Fig. 4 and Table S2), this is not the case in maize. Previous work has found that ∼10% of the ESTs sequenced from the shoot apical meristems of B73 and Mo17 were derived from retrotransposons (29), indicating they are transcriptionally active in stem cell populations at a level that could represent a metabolic cost to growth or rates of cell division. Maize differs from other plant species by containing a relatively high population of 22-nt siRNAs that does not depend on the RDR2/mop1 siRNA biogenesis pathway. Therefore, the impact of hybridization on sRNAs appears to depend on the content and organization of the genomes being investigated. We note that the genomes of many crop plants for which heterosis is important exhibit the complexity of repeats and paleoploidy characteristic of maize rather than minimal genomes of Arabidopsis or rice.
Based on their length and sequence similarity to the retrotransposon families from which they are derived, these siRNAs may act posttranscriptionally to degrade aberrant RNA transcribed from retrotransposons. If this were the case, we would expect an inverse relationship between mRNA levels and siRNAs for the retrotransposon families that differ between B73 and Mo17. When examined for the cinful retroelement, B73 has a higher abundance of cinful mRNA, 21-nt siRNAs, and 22-nt siRNAs compared with Mo17 (Fig. S3D). However, in the seedling shoot apex, we did see an increase in cinful expression as the shoot matured. This developmental shift corresponds to a decrease in miR156 (Fig. S3C), which, together with microRNA172 (miR172), controls phase change in maize (17, 30). It has been previously suggested in maize that a relaxation of TE silencing may be associated with vegetative phase change, so that the genome can recognize TEs and reinforce their silencing through sRNA pathways before reproductive development (31).
Our findings illustrate that the connection between rasiRNA levels and steady-state TE RNA levels is difficult to assess. First, assaying a single time point may not reveal down-regulation of TE RNA in rapidly growing tissues because TE silencing may be responding to developmental cues. Second, there may be a lag phase or possibly tissue preference to where down-regulation occurs. Because of the complexity of repetitive elements, feedback regulatory loops may exist that complicates the relationships between rasiRNA and target RNA levels, as has been documented for miR168 and AGO1 in Arabidopsis (24). Third, it is possible that the 21- to 22-nt siRNAs may have another purpose besides genome defense, and may possibly target genes. Ohtsu et al. (29) proposed that the expression of retrotransposons in dividing tissues may allow for the derivation of siRNAs that target genes with homologous sequences in their untranslated regions. Recently, direct evidence for this hypothesis has been obtained in Arabidopsis, in which a 21-nt siRNA derived from an Athila retrotransposon is produced in pollen cells and posttranscriptionally regulates the UBP1b gene that mediates stress response (28).
The relationship between retrotransposons and hybrid vigor is unclear, but the high degree of hybrid vigor displayed by maize compared with other plant species has been attributed to its highly repetitive genome (14). Genetic variation between parents is a requirement for hybrid vigor, and the two have a positive association on the average (25). East argued that the effects of hybrid vigor cannot be compared across genera because the relative degree of genetic differences likely varies (25). As we have demonstrated through sRNA sequencing, the retrotransposon portion of the maize genome provides an additional way for two inbred parents to differ by creating 21- to 22-nt populations of rasiRNAs. If TE-derived siRNAs can posttranscriptionally regulate endogenous genes as shown in Arabidopsis, the retrotransposon-derived 21- to 22-nt siRNAs in maize may serve as a significant source of regulatory variation acting at the posttranscriptional level. In a hybrid, the combination of divergent populations of 21- to 22-nt rasiRNAs may generate observed individual siRNA abundances falling between parental levels. However, given their putative function, new trans regulatory interactions mediated by these distinct 21- to 22-nt siRNAs could cause pleiotropic, developmentally dynamic, and synergistic molecular changes that contribute to the nonadditive phenotypic responses to hybridization in maize. Considering its immense genetic diversity and highly repetitive genome, genetic variation in a regulatory system mediated by TE-derived siRNAs could be a significant contributor to the dramatic vigor of maize hybrids.
Materials and Methods
Plant materials and experimental designs are described in SI Materials and Methods. Total RNA was isolated from plant tissues as described in the SI Materials and Methods. Library construction, sequencing, and analyses of sRNA sequences are described in SI Materials and Methods. The sRNA sequence data have been deposited in the Gene Expression Omnibus database.
Supplementary Material
Acknowledgments
The authors thank members of the M.E.H. and S.P.M. laboratories, especially Drs. Magdy Alabady, Qing Li, and Ying Li, for discussion. This work was supported by funds from the University of Illinois at Urbana–Champaign Critical Research Initiatives, National Science Foundation Grants 0922512 and 0501700 (to M.E.H. and S.P.M.) and US Department of Agriculture Cooperative State Research, Education, and Extension ServiceGrants 2004-35301-14495 (to J.E.D.) and 2011-67013-30041 (to S.P.M.). W.T.B. was supported by a William B. and Nancy L. Ambrose fellowship and a Bob Seifert Honorary Plant Breeding fellowship from Pioneer Hi-Bred.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE37411).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202073109/-/DCSupplemental.
References
- 1.Shull GH. The composition of field maize. American Breeding Association Reports. 1908;4:296–301. [Google Scholar]
- 2.Shull GH. What Is “Heterosis”? Genetics. 1948;33:439–446. doi: 10.1093/genetics/33.5.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 4.Messing J, Dooner HK. Organization and variability of the maize genome. Curr Opin Plant Biol. 2006;9:157–163. doi: 10.1016/j.pbi.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 5.Springer NM, et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 2009;5:e1000734. doi: 10.1371/journal.pgen.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 7.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha M, et al. Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA. 2009;106:17835–17840. doi: 10.1073/pnas.0907003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groszmann M, et al. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc Natl Acad Sci USA. 2011;108:2617–2622. doi: 10.1073/pnas.1019217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen H, et al. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell. 2012;24:875–892. doi: 10.1105/tpc.111.094870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He G, et al. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell. 2010;22:17–33. doi: 10.1105/tpc.109.072041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kenan-Eichler M, et al. Wheat hybridization and polyploidization results in deregulation of small RNAs. Genetics. 2011;188:263–272. doi: 10.1534/genetics.111.128348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birchler JA, Yao H, Chudalayandi S, Vaiman D, Veitia RA. Heterosis. Plant Cell. 2010;22:2105–2112. doi: 10.1105/tpc.110.076133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Springer NM, Stupar RM. Allelic variation and heterosis in maize: How do two halves make more than a whole? Genome Res. 2007;17:264–275. doi: 10.1101/gr.5347007. [DOI] [PubMed] [Google Scholar]
- 15.Nobuta K, et al. Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc Natl Acad Sci USA. 2008;105:14958–14963. doi: 10.1073/pnas.0808066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flint-Garcia SA, Buckler ES, Tiffin P, Ersoz E, Springer NM. Heterosis is prevalent for multiple traits in diverse maize germplasm. PLoS ONE. 2009;4:e7433. doi: 10.1371/journal.pone.0007433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, et al. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell. 2009;21:1053–1069. doi: 10.1105/tpc.109.065714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stupar RM, et al. Gene expression analyses in maize inbreds and hybrids with varying levels of heterosis. BMC Plant Biol. 2008;8:33. doi: 10.1186/1471-2229-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson C, et al. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19:1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voinnet O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 2008;13:317–328. doi: 10.1016/j.tplants.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 23.Dorweiler JE, et al. mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell. 2000;12:2101–2118. doi: 10.1105/tpc.12.11.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mallory AC, Vaucheret H. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 2009;10:521–526. doi: 10.1038/embor.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.East EM. Heterosis. Genetics. 1936;21:375–397. doi: 10.1093/genetics/21.4.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia Y, et al. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet. 2009;5:e1000737. doi: 10.1371/journal.pgen.1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shivaprasad PV, Dunn RM, Santos BA, Bassett A, Baulcombe DC. Extraordinary transgressive phenotypes of hybrid tomato are influenced by epigenetics and small silencing RNAs. EMBO J. 2011;31:257–266. doi: 10.1038/emboj.2011.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCue AD, Nuthikattu S, Reeder SH, Slotkin RK. Gene expression and stress response mediated by the epigenetic regulation of an Arabidopsis transposable element small RNA. PLoS Genet. 2012;8:e1002474. doi: 10.1371/journal.pgen.1002474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohtsu K, et al. Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.) Plant J. 2007;52:391–404. doi: 10.1111/j.1365-313X.2007.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Freeling M, Lisch D. Epigenetic reprogramming during vegetative phase change in maize. Proc Natl Acad Sci USA. 2010;107:22184–22189. doi: 10.1073/pnas.1016884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.