This work describes a unique role for the RNA-directed DNA methylation pathway (mainly CHH methylation, where H = A, T, or C) in mediating the parent-of-origin effect on the expression of the circadian clock gene CCA1 in Arabidopsis intraspecific hybrids. Altered CCA1 expression amplitudes are associated with heterosis of embryo growth and biomass accumulation in the reciprocal hybrids.
Abstract
Hybrid plants and animals often show increased levels of growth and fitness, a phenomenon known as hybrid vigor or heterosis. Circadian rhythms optimize physiology and metabolism in plants and animals. In plant hybrids and polyploids, expression changes of the genes within the circadian regulatory network, such as CIRCADIAN CLOCK ASSOCIATED1 (CCA1), lead to heterosis. However, the relationship between allelic CCA1 expression and heterosis has remained elusive. Here, we show a parent-of-origin effect on altered circadian rhythms and heterosis in Arabidopsis thaliana F1 hybrids. This parent-of-origin effect on biomass heterosis correlates with altered CCA1 expression amplitudes, which are associated with methylation levels of CHH (where H = A, T, or C) sites in the promoter region. The direction of rhythmic expression and hybrid vigor is reversed in reciprocal F1 crosses involving mutants that are defective in the RNA-directed DNA methylation pathway (argonaute4 and nuclear RNA polymerase D1a) but not in the maintenance methylation pathway (methyltransferase1 and decrease in DNA methylation1). This parent-of-origin effect on circadian regulation and heterosis is established during early embryogenesis and maintained throughout growth and development.
INTRODUCTION
Most living organisms have adapted to 24-h day/night cycles. Circadian rhythms are maintained by internal clocks and mediate physiology and metabolism in plants and animals (McClung, 2006; Wijnen and Young, 2006; Bass and Takahashi, 2010; Chen, 2010). In humans, energy intake and metabolism have a diurnal rhythm, and disturbance of the circadian rhythms leads to pathogenesis, including obesity, type 2 diabetes, and cardiovascular diseases (Prasai et al., 2008; Bass and Takahashi, 2010). clock−/− mutant mice lacking a diurnal feeding rhythm are hyperphagic and obese and develop a series of metabolic syndromes (Turek et al., 2005).
Accumulating evidence supports a role for the circadian clock in orchestrating carbohydrate metabolism, leading to hybrid vigor or heterosis. In Arabidopsis thaliana, circadian rhythms intrinsically regulate growth and development through diurnal changes in 30% or more of the transcriptome, including transcripts involved in photosynthesis and starch metabolism (Harmer et al., 2000; Smith et al., 2004; Covington et al., 2008). Transcriptional repressors including CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and their reciprocal regulators TIMING OF CAB EXPRESSION1 (TOC1) and GIGANTEA play critical roles in complex circadian regulatory networks in plants (McClung, 2006; Huang et al., 2012; Nagel and Kay, 2012). As a result, matching internal clock lengths with external light cycles increases CO2 fixation, growth, and fitness (Dodd et al., 2005). This is consistent with the recent finding of the circadian regulation of gene expression and photosynthetic activities in chloroplasts (Noordally et al., 2013). Indeed, altering the expression of circadian regulators increases chlorophyll biosynthesis and sugar and starch metabolism in Arabidopsis hybrids and allopolyploids (Ni et al., 2009). Hybrids can synthesize more starch during the day and degrade more starch at night. Clearly, the more sugar and starch that accumulates during the day, the more these molecules can be utilized through degradation at night (Graf et al., 2010) to promote growth. In the clock and starch mutants, starch cannot be fully degraded to promote growth, leading to growth retardation (Graf and Smith, 2011). Thus, this positive correlation of daytime starch accumulation with growth vigor and the negative association of morning residual starch levels with growth are not contradictory (Chen, 2013). The growth vigor in Arabidopsis hybrids and allotetraploids is partly controlled by the epigenetic regulation of central circadian clock oscillator components, including CCA1 (Ni et al., 2009). Notably, the association of increased biomass with CCA1 repression during the day has been independently demonstrated in Arabidopsis intraspecific hybrids (Miller et al., 2012; Shen et al., 2012). In superhybrid rice (Oryza sativa), yield-related quantitative trait loci are also associated with gene expression changes in the circadian clock and light signaling pathways (Song et al., 2010). Moreover, down-regulation of CCA1 expression during the day in Arabidopsis diploids leads to more starch at dusk and increased biomass, a phenotype similar to that seen in the hybrids (Ni et al., 2009).
The mechanism for regulating maternal and paternal alleles of clock genes that affect heterosis remains unknown. In Arabidopsis, de novo methylation of CHH and CHG sites (where H = A, T, or C) is established through the RNA-directed DNA methylation (RdDM) pathway (Wassenegger et al., 1994; Aufsatz et al., 2002). NUCLEAR RNA POLYMERASE D1a (NRPD1a) encodes a large subunit of RNA polymerase IV, which is essential for the biogenesis of 24-nucleotide small interfering RNAs (siRNAs) (Herr et al., 2005; Onodera et al., 2005). Double-stranded siRNAs are amplified through RNA-DEPENDENT RNA POLYMERASE2 and cleaved by the endoribonuclease DICER-LIKE3. These siRNAs are then loaded onto ARGONAUTE4 (AGO4), and AGO4-associated siRNAs are predicted to guide the de novo methyltransferase activity of DOMAINS REARRANGED METHYLASE2 (Zilberman et al., 2004; Gao et al., 2010), leading to RdDM (Haag and Pikaard, 2011; Law et al., 2013). The maintenance of DNA methylation requires METHYLTRANSFERASE1 (MET1), which encodes a DNA methyltransferase, as well as DECREASE IN DNA METHYLATION1 (DDM1), which is a SWI/SNF2-like chromatin-remodeling protein (Jeddeloh et al., 1999). DNA methylation affects the expression of circadian clock genes. For example, CCA1 and LHY were upregulated in Arabidopsis DNA methylation mutants (Zhang et al., 2006; Kurihara et al., 2008) and in plants treated with 5′-aza-2′-deoxycytidine, which inhibits DNA methylation (Shen et al., 2012).
Changes in CCA1 expression and DNA methylation were observed in Arabidopsis hybrids (Shen et al., 2012), but the relationship between them is unclear. The parent-of-origin effect on biomass vigor in reciprocal Arabidopsis hybrids suggests an epigenetic cause (Miller et al., 2012). In allotetraploids derived from Arabidopsis and Arabidopsis arenosa, the maternally transmitted Arabidopsis CCA1 allele is more repressed than the paternally transmitted A. arenosa allele, which is associated with histone modifications (Ni et al., 2009). Here, we tested the hypothesis that the altered expression of circadian genes in hybrids is mediated by epigenetic factors such as DNA methylation (Chen, 2013). We investigated how and when the parent-of-origin effect on CCA1 expression and growth vigor is established in Arabidopsis hybrids. The results support that NRPD1a- and AGO4-mediated changes in CHH methylation affect CCA1 expression amplitudes and growth vigor in hybrids during vegetative and embryo development.
RESULTS AND DISCUSSION
Parent-of-Origin Effects on Starch and Biomass Heterosis in Hybrids
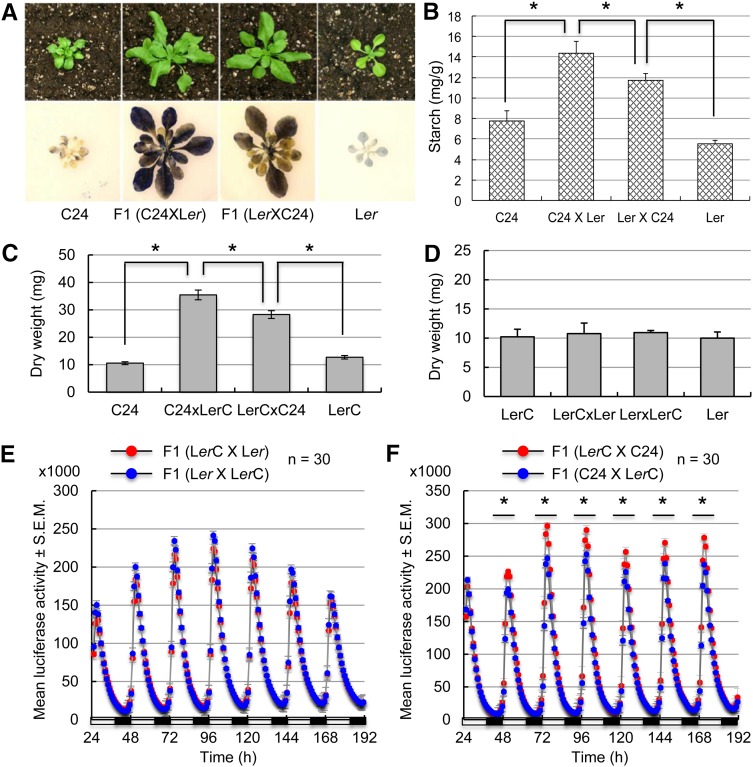
Biomass heterosis has been obviously observed in reciprocal F1 hybrids between Arabidopsis C24 and Landsberg erecta (Ler) (Meyer et al., 2004; Groszmann et al., 2011; Shen et al., 2012). The reciprocal hybrids accumulated 50 and 100% more starch in rosette leaves at dusk (ZT15; Zeitgeber time 0 [ZT0] = dawn) than their parents C24 and Ler (Figure 1A). Interestingly, the starch content was ∼22% higher in the F1 (C24 × Ler) hybrid (by convention, the maternal parent is listed prior to the paternal parent) than in the reciprocal F1 (Ler × C24) hybrid (P < 0.05, Student’s t test) (Figure 1B). The biomass (dry weight) was also significantly higher when C24 was the maternal parent in the reciprocal crosses (Figure 1C; P < 0.05). This parent-of-origin effect on biomass vigor was also observed in other reciprocal F1 hybrids between Columbia-0 (Col-0) and C24 or between Col-0 and Ler (Miller et al., 2012), although the vigor level was low in the latter. The variable degree of biomass vigor among different hybrids may reflect genotypic effects (Meyer et al., 2004; Chen, 2010). Collectively, these data suggest a parent-of-origin effect on biomass heterosis. To minimize potential genotypic effects, unless noted otherwise, further analyses were performed in F1 crosses between C24 and Ler ecotypes, including several mutants in the Ler background.
Figure 1.
Parent-of-Origin Effects on CCA1 Expression and Biomass Accumulation in F1 Hybrids.
(A) Larger size (top row) and higher starch-staining intensities at ZT15 (bottom row) in F1 (C24 × Ler) than in F1 (Ler × C24).
(B) Quantification of starch content at ZT15 in C24, Ler, and C24 × Ler and Ler × C24 reciprocal F1 hybrids. Asterisks indicate a statistical significance level of 0.05 (Student’s t test).
(C) Higher aerial dry weight ± sd (y axis) of 3-week-old seedlings in F1 (C24 × LerC) than in F1 (LerC × C24), relative to the parents C24 and LerC (P < 0.05; n = 5 in each of three replicates).
(D) No difference in aerial dry weight (y axis) in 3-week-old seedlings of reciprocal crosses between Ler and LerC (n = 5 in each of three replicates).
(E) Mean values ± se of bioluminescence counts (y axis; in thousands) in seedlings of the reciprocal F1 crosses LerC × Ler (red) and Ler × LerC (blue). Each data point was averaged from 24 plants with se. The x axis shows hours with an alternating cycle of light (open bars) and dark (closed bars).
(F) Mean values ± se of bioluminescence counts (y axis; in thousands) in seedlings of the reciprocal hybrids LerC × C24 (red) and C24 × LerC (blue). Specifics are as in (E). Black lines with asterisks indicate the range of time points with statistically significant differences between the reciprocal crosses (P < 0.05, Student’s t test).
Parent-of-Origin Effects on Circadian Rhythms
Altered CCA1 expression correlated with growth vigor in allopolyploids, hybrids, and diploids (Ni et al., 2009). Repressing CCA1 peaks in TOC1:cca1(RNAi) transgenic plants during the day increases starch content and biomass, while overexpressing TOC1:CCA1 in transgenic plants decreases starch content and biomass. These data suggest an important role for altered CCA1 expression amplitudes in promoting growth vigor. In the hybrids between C24 and Ler, expression levels of endogenous CCA1 were 20 to 30% lower than the midparent value at ZT6 (P < 0.05) (Supplemental Figure 1A). RT-PCR analysis showed that CCA1 expression peaks were lower in the cross when C24 was the maternal parent than in the reciprocal cross at ZT6, and lower CCA1 expression levels were correlated with higher levels of starch and biomass in C24 × Ler hybrids when C24 was the maternal parent (Figures 1B and 1C), suggesting an anticorrelation between endogenous CCA1 expression levels and biomass vigor in hybrids.
RT-PCR analysis is limited to specific time points when the tissues can be collected for diurnal analysis. To overcome this caveat, we employed a stable Ler (CCA1:LUC or ProCCA1:LUC) transgenic line (Salomé and McClung, 2005), which we designated LerC (Figure 1E; Supplemental Figures 1B and 1C). Using the reporter line, we tested how diurnal oscillation of CCA1 expression in a period of 5 to 7 d is associated with biomass vigor in hybrids. Unless noted otherwise, bioluminescence assays included three biological replicates each with 24 to 32 seedlings, and resulting data points at ∼1-h intervals were analyzed for statistical significance using paired Student’s t tests between each comparison (e.g., reciprocal hybrids) (see Methods). In the control crosses, similar biomass levels (Figure 1D; Supplemental Figure 1B) correlated with equal expression levels of CCA1:LUC through the paternal or the maternal parent in reciprocal crosses between Ler and LerC lines (Figure 1E). In reciprocal F1 hybrids between C24 and LerC (Supplemental Figure 1C), the CCA1 expression peak was statistically significantly lower when CCA1:LUC was transmitted through the paternal parent (in C24 × LerC) than the maternal parent (in LerC × C24) (Figure 1F) (P < 0.05, Student’s t test). Consistently, lower CCA1 expression peaks in the C24 × LerC cross correlated with higher starch and biomass levels in the C24 × LerC hybrids than in the reciprocal cross (Figures 1B and 1C). These data indicate a parent-of-origin effect on the expression of transgene CCA1:LUC and endogenous CCA1, which negatively correlates with biomass heterosis.
CHH Methylation and AGO4 Affect Parent-of-Origin Effects on Circadian Gene Expression
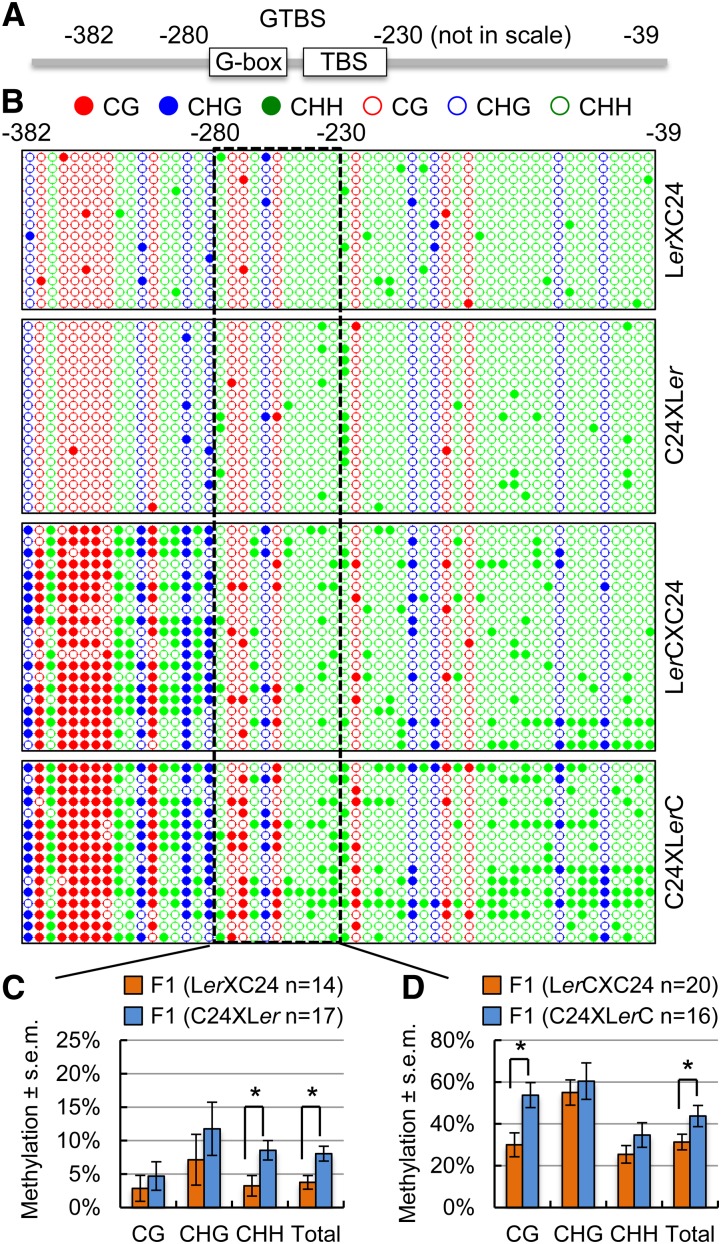
Parent-of-origin effects are often associated with paternal and maternal inheritance of DNA methylation patterns (Huh et al., 2008; Ferguson-Smith, 2011; Raissig et al., 2011), and methylation levels in promoter regions correlate with gene expression levels (Zilberman et al., 2007). To test this, we examined the methylation levels of CG, CHG, and CHH (where H = A, T, or C) sites in the CCA1 promoter region using the bisulfite sequencing method (Gruntman et al., 2008). Degenerate primers flanking the CCA1 promoter region (−382 to −39, relative to the transcription start site of +1) were used to amplify bisulfite-treated DNA, which was subsequently cloned and sequenced. Methylation levels of CHH, CHG, and CG sites were calculated and compared between reciprocal hybrids. In addition to this larger promoter fragment (−382 to −39), methylation analyses were also performed on a smaller region including a motif domain (−280 to −230) that contains a G-box and a CCA1 HIKING EXPEDITION (CHE), a class I TCP protein, binding site (Pruneda-Paz et al., 2009), which is named GTBS (Figure 2A; Supplemental Figure 2A). This GTBS region was selected for the methylation analysis because CHE binds there to mediate CCA1 expression (Pruneda-Paz et al., 2009). In reciprocal F1 hybrids between C24 and Ler (wild type), within the GTBS region, CHH methylation levels were statistically significantly higher in C24 × Ler hybrids than in Ler × C24 hybrids (Figures 2B and 2C) (P < 0.05, Student’s t test). CG and CHG methylation levels showed large variation, and this variability could result from a few CG and CHG sites, compared with CHH sites, in the promoter regions analyzed (Figure 2B). The differences in CHH methylation in the GTBS region between the reciprocal F1 hybrids of C24 and Ler were correlated negatively with the endogenous CCA1 expression and positively with biomass and starch content in F1 hybrids between C24 and Ler (Figure 1B; Supplemental Figure 1A). In the larger CCA1 promoter region, methylation levels of CHH sites were also higher in the C24 × Ler hybrids than in the Ler × C24 hybrids, although such differences were statistically insignificant (Supplemental Figure 1D).
Figure 2.
Bisulfite Sequencing Analysis of DNA Methylation in Reciprocal Hybrids.
(A) Diagram of a larger promoter region (−382 to −39) of CCA1 and a motif region (−280 to −230) that contains a G-box and CHE with a class I TCP protein binding site, named GTBS.
(B) Dot-plot analysis of CG, CHG, and CHH methylation changes in Ler × C24 (top), C24 × Ler (second), LerC × C24 (third), and C24 × LerC (bottom) in the large promoter region and the GTBS region (boxed). A total of 14 to 20 individual promoter fragments were sequenced and analyzed in each sample. Red, blue, and green circles indicate CG, CHG, and CHH methylation (closed) or no methylation (open).
(C) Percentage of methylation changes ± se in the GTBS region between reciprocal hybrids of Ler × C24 (orange) and C24 × Ler (blue). n = number of clones sequenced in each replicate. Asterisks indicate a statistical significance level of 0.05 (Student’s t test).
(D) Percentage of methylation changes ± se in the GTBS region between reciprocal hybrids of LerC × C24 (orange) and C24 × LerC (blue). n = number of clones sequenced in each replicate. Asterisks indicate a statistical significance level of 0.05 (Student’s t test).
We further analyzed DNA methylation in reciprocal F1 plants between LerC (transgenic line) and C24 or Ler (Figures 2B and 2D; Supplemental Figures 1E and 2). In the hybrids between C24 and LerC, methylation levels at CHH sites in the larger CCA1 promoter region were significantly higher in C24 × LerC hybrids when C24 was the maternal parent than in LerC × C24 hybrids (Supplemental Figure 2C). Within the GTBS, the trend of higher CHH methylation in C24 × LerC hybrids remained, but the difference between the reciprocal crosses was statistically insignificant (Figure 2D). However, total and CG methylation levels were statistically significantly higher in the C24 × LerC cross than in the reciprocal cross (Figure 2D). This is probably because of an increased methylation level in the CG sites of the transgene promoter (Supplemental Figure 3), which may obscure the difference in CHH methylation. In control crosses between LerC and Ler, methylation levels of all sites were similar in the larger CCA1 promoter region (Supplemental Figures 2B and 2D) or within the GTBS (Supplemental Figure 1E).
Higher methylation levels in the transgenic CCA1:LUC locus than in the endogenous CCA1 locus were also observed in the 5′ untranslated region (UTR) (Supplemental Figure 3A), in which endogenous and transgene loci could be discriminated in the F1 hybrids (Supplemental Figures 3B and 3C). However, the methylation level differences between the reciprocal hybrids were not significant (Supplemental Figures 3D to 3F). These data suggest that the transgene is highly methylated, especially at CG and CHG sites, and that the methylation changes in the 5′ UTR are not correlated with the transgene or endogenous CCA1 expression in the reciprocal hybrids. Instead, CHH methylation changes in the GTBS region are correlated with lower CCA1 expression when C24 is the maternal parent in the hybrids.
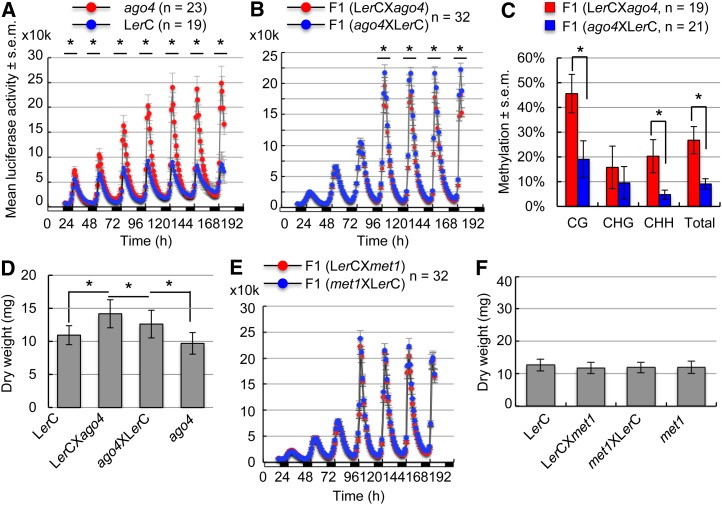
In the RdDM pathway (Law and Jacobsen, 2010; Haag and Pikaard, 2011), AGO4 controls locus-specific methylation of CHH and CHG sites (Zilberman et al., 2004; Gao et al., 2010). We first tested if CCA1:LUC expression is altered in the ago4-1 mutant. Indeed, CCA1:LUC expression levels were statistically significantly higher in the ago4-1 homozygous mutant (red) than in the wild type (blue) (Figure 3A) (P < 0.05, Student’s t test). We then tested if CCA1:LUC expression is altered in reciprocal F1 crosses involving the ago4-1 mutant. Remarkably, in the F1 crosses between the ago4-1 (Ler) mutant and LerC, the CCA1:LUC expression peak was statistically significantly higher when the ago4-1 mutation was carried on the maternal genome (blue) than on the paternal genome (red) (Figure 3B), suggesting gene silencing by the RdDM pathway (Wassenegger et al., 1994; Aufsatz et al., 2002) through the action of maternal siRNAs in leaves as in the endosperm (Mosher et al., 2009; Lu et al., 2012). These siRNAs induce RdDM (Law and Jacobsen, 2010; Haag and Pikaard, 2011), and consistent with this notion, the methylation levels of all sites were lower in the ago4-1 × LerC cross than in the LerC × ago4-1 cross in both the larger promoter region (P < 0.05, Student’s t test) (Supplemental Figure 5A) and the GTBS region (Figure 3C). The biomass was significantly higher in the LerC × ago4-1 cross than in the ago4-1 × LerC cross (P = 0.05, Student’s t test) (Figure 3D; Supplemental Figure 6A). In reciprocal F1 hybrids between C24 and ago4-1 (Ler), the biomass in F1 (ago4-1 × C24) was increased, making it similar to but not higher than that in F1 (C24 × ago4-1) (Supplemental Figures 6C and 6E). Notably, siRNAs and RdDM pathways may also regulate other traits, such as seed size. For example, seed size was dramatically increased when the maternal siRNAs were reduced (Lu et al., 2012) or when the demethylation lines were the maternal parent in the genetic crosses (Adams et al., 2000; Xiao et al., 2006). In addition, biomass accumulation is affected by other factors, such as quantitative trait loci, that regulate metabolism during the early stages of seedling development (Lisec et al., 2009; Meyer et al., 2012).
Figure 3.
Parent-of-Origin Effects on CCA1 Expression Depend on CHH Methylation and AGO4 in Reciprocal Hybrids.
(A) Mean values ± se of bioluminescence counts (in ten thousands [10k]; y axis) for CCA1:LUC expression in seedlings of the wild-type (LerC) (blue) and the ago4-1 homozygous mutant (ago4) (red). Each data point was averaged from 19 and 23 plants, respectively. Black lines with asterisks indicate the peak time points with statistically significant differences between the reciprocal F1 crosses (P < 0.05, Student’s t test). The x axis shows hours with an alternating cycle of light (open bars) and dark (closed bars).
(B) Mean values ± se of bioluminescence counts (in ten thousands [10k]; y axis) in seedlings of the reciprocal F1 crosses between LerC × ago4-1 (red) and ago4-1 × LerC (blue). Specifics are as in (A).
(C) Percentage of methylation changes in the GTBS region between reciprocal F1 crosses of LerC × ago4-1 (red) and ago4-1 × LerC (blue). Asterisks indicate statistical significance (P < 0.05, Student’s t test).
(D) Aerial dry weight comparison (y axis) between 3-week-old seedlings in LerC × ago4-1 and in ago4-1 × LerC crosses (n = 15; P = 0.05, Student’s t test) and between the reciprocal F1 seedlings and the ago4-1 or LerC parent (P < 0.05).
(E) Mean values ± se of bioluminescence counts (in ten thousands [10k]; y axis) in seedlings of the reciprocal F1 crosses between LerC × met1-1 (red) and met1-1 × LerC (blue). Each data point was averaged from 32 plants with se.
(F) No difference in aerial dry weight (y axis) in 3-week-old seedlings of reciprocal F1 crosses between met1-1 and LerC (P = 0.7, Student’s t test).
Consistent with the role for the RdDM pathway in circadian gene expression, the expression direction of CCA1:LUC was also altered in the reciprocal F1 crosses ColC (Col-0 [CCA1:LUC or ProCCA1:LUC] transgenic line) × nrpd1a and nrpd1a × ColC (Supplemental Figure 7B), whereas CCA1:LUC was equally expressed in the control crosses (Supplemental Figure 7A). Note that Col-0 was used in this study because the nrpd1a mutant and ColC or Col-0 (ProCCA1:LUC) would be in the same genetic background. The altered direction of expression was different from the crosses involving ago4-1, which could be associated with different genotypes (Col-0 versus Ler) and/or different steps of NRPD1a and AGO4 involved in the RdDM pathway (Law and Jacobsen, 2010; Haag and Pikaard, 2011).
To test if the maintenance of DNA methylation affects CCA1 expression, we made reciprocal F1 crosses between LerC and ddm1-2 (Ler) (Jeddeloh et al., 1998) or met1-1 (Ler) (Kankel et al., 2003). In the met1-1 mutant, CCA1:LUC expression remained unchanged relative to that in the wild type (Supplemental Figure 5C). Consequently, no alteration of the parent-of-origin effect on CCA1 expression amplitudes was found in F1 crosses between LerC and the met1-1 mutant (Figure 3E), which had similar biomass (Figure 3F; Supplemental Figure 5D). Similarly, in the reciprocal crosses between LerC and the ddm1-2 mutant, CCA1 expression amplitudes were equal in the F1 crosses regardless of whether LerC was used as the maternal or the paternal parent (Supplemental Figure 7C). In reciprocal F1 crosses between C24 and ddm1-2, the parent-of-origin effect on biomass remained unchanged, namely, higher in C24 × ddm1-2 than in ddm1-2 × C24 (Supplemental Figures 6D and 6F). Methylation analysis showed that CG and CHG methylation levels but not CHH methylation were lower in ddm1-2 × LerC than in LerC × ddm1-2 in the larger promoter fragment (Supplemental Figure 5B) or in the GTBS region (Supplemental Figure 7D). However, only CG methylation was significantly different (P < 0.05, Student’s t test) between the reciprocal F1 plants, and the direction for the parent-of-origin effect on CCA1:LUC expression remained unchanged. These data suggest that DDM1 and MET1 affect mainly CG and CHG methylation but do not alter CHH methylation or parent-of-origin effects on CCA1 expression and biomass.
Parent-of-Origin Effects on CCA1 Expression during Early Stages of Embryo Development
A key question is, when is the parent-of-origin effect on circadian rhythms established? Plant cells are totipotent and contain cell-autonomous multiple-loop clocks (McClung, 2006; Harmer, 2009; Nagel and Kay, 2012). The circadian clock is obviously present in leaves but also in roots and shoots (James et al., 2008) and germinating seeds (Penfield and Hall, 2009). This suggests that the clock may function during embryo and seed development and that preferential expression of the maternal CCA1 allele may be established during these stages. We first examined the expression of CCA1, LHY, TOC1, and CHE in developing siliques 5 d after pollination in two ecotypes (Ler and Col-0). The mRNA abundance of two morning-phased genes, CCA1 and LHY, peaked at ZT0, rapidly decreased toward a minimum at ZT12, and then rapidly increased toward ZT24 (Supplemental Figures 8A to 8D). The evening-phased genes, TOC1 and CHE, exhibited antiphasic diurnal expression patterns compared with those of CCA1 and LHY (Supplemental Figures 8E to 8H). These data suggest that a robust clock is maintained in developing siliques as in leaves, roots (James et al., 2008), and germinating seeds (Penfield and Hall, 2009).
In addition to an embryo, a typical seed contains an endosperm and a seed coat, which are maternal tissues that do not transmit genetic information to the next generation. To test if the clock is functional in developing embryos, we dissected embryos 10 d after pollination (see Methods), when the embryos could grow in culture medium (Figures 4A to 4C; Supplemental Figures 9A and 9B). After 4 d on culture medium, embryos were subjected to bioluminescence assays (Figure 4D; Supplemental Figures 9C to 9E). Consistent with circadian rhythms in seedling leaves, CCA1:LUC expression amplitudes were statistically significantly higher when the transgene was transmitted through the maternal parent (in LerC × C24) than through the paternal parent (in C24 × LerC) (Figure 4D), which correlated negatively with embryo growth rates (Figure 4C). In control crosses between LerC and Ler, the embryos were of similar size (Supplemental Figure 9A), and no CCA1:LUC expression difference was observed in the embryos of reciprocal F1 hybrids (Supplemental Figure 9C). The stronger maternal expression of CCA1 was also found in the embryos of another pair of reciprocal F1 hybrids between Ler and C24 (CCA1:LUC) or the C24 (CCA1:LUC or ProCCA1:LUC) transgenic line (C24C) (Supplemental Figures 9B and 9E), while in embryos of the control crosses, CCA1:LUC expression levels were equal (Supplemental Figure 9D).
Figure 4.
Parent-of-Origin Effect on Circadian Rhythms and Embryo Growth Rates in Hybrids.
(A) and (B) Embryos of C24 × LerC (A) and LerC × C24 (B) were dissected 10 d after pollination and cultured in medium for 1 d (day 1). After 4 d (day 4) in culture, C24 × LerC (A) and LerC × C24 (B) embryos were subjected to bioluminescence assays. Bars = 0.1 mm.
(C) Changes in embryo growth rates between C24 × LerC (blue) and LerC × C24 (red) crosses were determined by measuring embryo surface areas (means ± sd) (y axis) during 10 d in culture (x axis). n = 5 in each of three replicates.
(D) Mean values ± se of bioluminescence counts (y axis) in cultured embryos of the reciprocal F1 hybrids LerC × C24 (red) and C24 × LerC (blue).
A Model for Parent-of-Origin Effects on Altered Circadian Rhythms and Growth Vigor in Hybrids
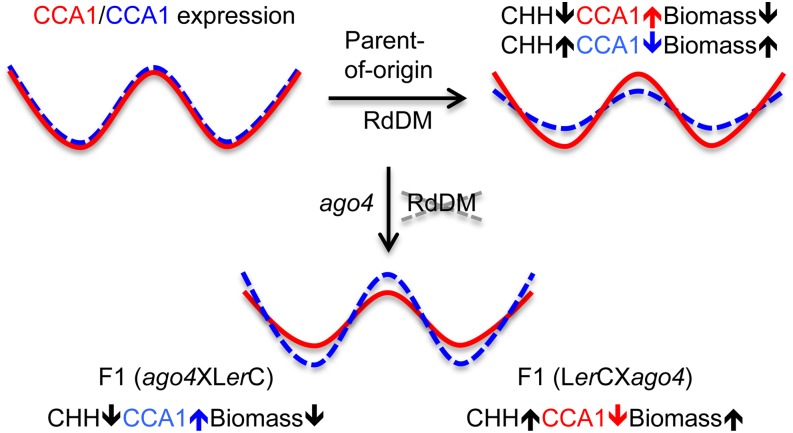
The available data support a model that explains how changes in CCA1 expression mediate growth vigor in hybrids (Figure 5). Changes in circadian expression amplitude (or phase) without altering the clock period can have significant consequences for clock-controlled metabolic rhythms in animals (Nakahata et al., 2009) as well as in plants (Ni et al., 2009). Epigenetic repression of the maternal CCA1 allele is correlated with increased starch content and biomass in Arabidopsis hybrids and allotetraploids (Ni et al., 2009). Correlation of CCA1 repression with growth vigor is confirmed in Arabidopsis hybrids (Miller et al., 2012; Shen et al., 2012). This repression of the CCA1 expression peak is established during embryo development, which requires AGO4 and other components such as NRPD1a in RdDM (Gao et al., 2010; Law and Jacobsen, 2010). Quantitative variation of CHH methylation levels in the regulatory motif (GTBS) of the CCA1 promoter region correlates with CCA1 expression amplitudes and depends on the NRPD1a- and AGO4-mediated pathway, possibly through interactions with 24-nucleotide siRNAs. There is evidence for maternal transmission of 24-nucleotide siRNAs in endosperm (Mosher et al., 2009; Lu et al., 2012) and an overall increase of 24-nucleotide siRNAs in Arabidopsis hybrids relative to the parents (Groszmann et al., 2011; Shen et al., 2012). These data suggest a role for siRNAs in the parent-of-origin effect on CCA1 expression. AGO4 could recognize maternal and paternal siRNAs and guide parent-of-origin effects on DNA methylation. This discrimination mechanism may not depend solely on the primary DNA sequence, because promoter sequences between two ecotypes are the same. The promoters could be associated with differential modifications of chromatin, including histone acetylation and methylation. For example, H3K27me3 could induce CHG methylation through the action of CMT3 (Cao et al., 2003) or CHH methylation through the action of CMT2 (Zemach et al., 2013). As a result, methylation of the CCA1 promoter region inhibits the binding of CHE and other proteins to GTBS, altering CCA1 expression. When the CHH methylation level in the promoter is high, CCA1 is repressed and biomass is increased (Figure 5, right). Disruption of AGO4 in the F1 crosses reverses CHH methylation levels in the promoter of paternal and maternal CCA1 alleles and their expression directions, leading to altered biomass accumulation (Figure 5, bottom). When CHH methylation levels in the promoter are reduced, the paternal CCA1 is increased and biomass is also decreased (Figure 5, bottom left). AGO4 is required for changes in CHH methylation but not sufficient to alter biomass vigor at a statistically significant level. In plants, biomass vigor could be affected by many other epigenetic factors and metabolic pathways (Chen, 2013). For example, disruption of the RdDM pathway could alter the expression of other genes in stress response and metabolic pathways, which in turn alter growth vigor. The opposite is true in the reciprocal cross (Figure 5, bottom right). However, disruption of the maintenance of DNA methylation (mainly CG) through ddm1 or met1 mutations does not change the parent-of-origin effect on CCA1 expression and biomass.
Figure 5.
A Model for the Parent-of-Origin Effect on Circadian Rhythms and Growth Vigor in Hybrids.
At top left, maternal (red, solid line) and paternal (blue, dashed line) CCA1 alleles are equally expressed in reciprocal crosses in the same genotype. At top right, in reciprocal F1 hybrids, when CHH methylation levels are low in the promoter of the maternal CCA1 allele, its expression is high, whereas when CHH methylation levels are high in the promoter of the paternal CCA1 allele, its expression level is low. This parent-of-origin effect on CCA1 expression is anticorrelated with biomass accumulation. At bottom, in F1 crosses involving the ago4 mutant, disruption of RdDM leads to lower CHH methylation levels in the promoter of the paternal allele than that of the maternal allele. As a result, paternal CCA1 expression is increased and biomass is decreased (left). In the reciprocal cross, maternal CCA1 expression is lower than that of the paternal allele (right). The reversal of the parent-of-origin effect on CCA1 expression leads to increased levels of biomass accumulation.
Methylation and small RNA changes were previously observed in the same hybrids between C24 and Ler (Groszmann et al., 2011; Shen et al., 2012), but correlation of CCA1 and LHY repression with specific methylation sites was not obvious. It is possible that genome-wide assays could not provide in-depth analysis of these loci in specific regions because the read coverage ranges from 2 to 47% of the genome (Shen et al., 2012). We predict that siRNA and RdDM pathways may also affect other targets or regulators such as LHY, TOC1, and CHE in the circadian feedback loop and related networks, which in turn mediate CCA1 expression (McClung, 2006; Harmer, 2009; Nagel and Kay, 2012). Reduction in CCA1 expression amplitudes or transcript levels promotes the expression of downstream genes that are negatively regulated by CCA1 abundance, as shown in Arabidopsis diploids and allopolyploids (Ni et al., 2009). This altered circadian regulation could affect photosynthetic and metabolic pathways that are altered in F1 hybrids (Fujimoto et al., 2012; Meyer et al., 2012) as well as overall regulatory networks related to growth and development (Birchler et al., 2010).
This model of the parent-of-origin effect on circadian rhythms and growth vigor is consistent with the parental conflict theory for imprinting in mammals and flowering plants, which predicts that the maternal genome provides factors that inhibit growth whereas the paternal genome carries the factors that promote growth (Moore and Haig, 1991; Ferguson-Smith, 2011; Raissig et al., 2011; Haig, 2013). This parental conflict theory could apply to the maternal effect of the clock function on growth vigor during the early stages of embryo development in hybrids and sexually reproducing organisms. When the maternal CCA1 is repressed, growth vigor is increased. When the maternal CCA1 expression is upregulated, growth vigor is reduced. The parent-of-origin effect on an early mouse embryo phenotype was also reported (Han et al., 2008). This parent-of-origin effect on circadian rhythms and growth vigor in embryos is likely a general phenomenon to regulate growth and development in plant hybrids and allopolyploids as well as in sexually reproducing organisms including mammals.
METHODS
Plant Materials
Plant materials included three Arabidopsis thaliana ecotypes, C24, Ler, and Col-0, three mutants in DNA methylation genes, met1-1 (Ler) (Kankel et al., 2003), ddm1-2 (Ler) (Jeddeloh et al., 1998), and ago4-1 (CS6364; Ler), and a small RNA biogenesis mutant, nprd1a-4 (CS66151). met1-1 and ddm1-2 mutant seeds were kindly provided by Eric Richards at the Boyce Thompson Institute for Plant Research, and ago4-1 and nprd1a-4 were obtained from the ABRC. For comparison in F1 hybrids and crosses, manual pollination was used to produce seeds in both parents and reciprocal hybrids. For gene expression and starch analyses in vegetative tissues, plants were grown for 3 weeks in 16/8-h light/dark cycles at 22/18°C and harvested at ZT0 (dawn). Rosette leaves were harvested from a pool of 6 to 12 plants as one biological replicate and used immediately or frozen in liquid nitrogen for future use. Leaves were collected prior to bolting to minimize developmental variation among genotypes. Except where noted otherwise, three replicates were used for each experiment.
For gene expression analysis in developing siliques, manual pollination was performed 1 d after emasculation. Young siliques at 5 d after pollination were harvested every 3 h for a period of 24 h for diurnal expression analysis.
Transgenic Plants Expressing Luciferase Reporter
The ProCCA1:LUC construct was transformed into Ler as described previously (Salomé and McClung, 2005) to generate LerC stable transgenic plants for this study. To generate the ColC and C24C lines (Supplemental Figures 7 and 9), a CCA1 promoter (from −715 to −1 bp, relative to the transcription start site plus full 5′ UTR) was amplified by PCR and cloned into the plasmid between the restriction sites XhoI and NcoI. A ProCCA1:LUC plasmid construct was generated by inserting the luciferase gene between the restriction enzyme sites NcoI and BamHI in the pFAMIR plasmid that was modified from pFGC5941 (McGinnis et al., 2005). The construct was introduced into Arabidopsis (Col-0 or C24) plants using Agrobacterium tumefaciens–mediated transformation (strain GV3101) with the floral dip method (Clough and Bent, 1998). Primary transformants (seedlings) were screened on Murashige and Skoog (MS) agar medium (M9274; Sigma-Aldrich) (Murashige and Skoog, 1962) supplemented with 7.5 μg/mL Basta (Sigma-Aldrich). Stable transgenic plants (T2 and later) with uniform herbicide resistance were used for the expression assays and for making crosses. For hybrid crosses involving Ler and C24 ecotypes, either the LerC or C24C reporter line was used. For crosses involving the nrpd1a mutant, ColC was used such that the mutant and the reporter lines are in the same ecotype background.
Embryo Dissection and Culture
Siliques at 10 d after pollination were harvested and rinsed with 70% ethanol and soaked in 100% Clorox for 2 min. After rinsing with autoclaved water twice, the siliques were kept in sterilized liquid MS medium (Murashige and Skoog, 1962) in a Petri dish. Embryos were dissected using an optical microscope (SMZ445; Nikon) and transferred to a plate containing the agar embryo culture medium, which contained 40% Suc, 0.5× MS salts, 0.9 mg/L thiamine, 0.5 g/L MES (Sigma-Aldrich), 8 g/L agar, and 0.69 g of Leu-DO amino acid supplements (Clontech). Final pH was adjusted to 5.9 with KOH. Forty to 50 embryos from each genotype were transferred to one agar plate and cultured in an incubator at 22°C (16/8-h light/dark cycles) for 2 d. A total of 24 healthy embryos (no brown spots or any visible damage) from each genotype were transferred to a 96-well microtiter plate (Nagle Nunc International) containing 40% Suc, 0.5× MS salts (Murashige and Skoog, 1962), 0.9 mg/L thiamine, 0.5 g/L MES (Sigma-Aldrich), 8 g/L agar, and 0.69 g of Leu-DO amino acid supplements. After adding luciferin to a final concentration of 2.5 mM, the plate was subjected to luciferase assays over a period of 5 to 7 d (see below).
Luciferase Assays and Data Analysis
Stable transgenic embryos or seedlings containing ProCCA1:LUC constructs were analyzed using a TopCount NXT luminometer and scintillation counter (Perkin-Elmer). For seedlings, seeds were sterilized and plated on 1% (w/v) agar MS medium (Murashige and Skoog, 1962) plus 30 g/L Suc. Seeds were stratified for 2 d in the dark at 4°C and then transferred to a 16-h-light/8-h-dark cycle for 8 d at 22°C. Seedlings were transferred to white microtiter plates (Nagle Nunc International) containing agar MS medium plus 30 g/L Suc, and then 30 μL of 0.5 mM luciferin (Gold Biotechnology) was added to each well. Microtiter plates were covered with clear plastic MicroAmp sealing film (Applied Biosystems), in which holes were placed above each well for seedling gas exchange. Plates were moved to the TopCount device and interleaved with three clear plates to allow light diffusion to the seedlings. Luciferase activity was measured approximately every 1 h by integrating photons emitted by seedlings during a 10-s sampling period. Data were analyzed by fast Fourier transform-nonlinear least squares (Plautz et al., 1997) using the Biological Rhythms Analysis Software System Excel macros (available from http://millar.bio.ed.ac.uk/PEBrown/BRASS/BrassPage.htm).
All values are presented as means ± se. Expression amplitudes are shown as bioluminescence counts. Unless noted otherwise, each data point was averaged from 24 to 32 plants in each experiment, and graphic data from one of three replicated experiments are shown.
RNA Preparation and RT-PCR
Total RNA was extracted using Plant RNA reagent (Invitrogen). First-strand cDNA synthesis was performed using RT SuperScript III (Invitrogen). For RT-PCR, the total RNA obtained from siliques was treated with RNase-free DNase (Promega) for 30 min. The reaction was terminated by adding phenol:chloroform:isoamyl alcohol (25:24:1) solution, and the total RNA was precipitated by the addition of ethanol. An aliquot (1:100) of cDNA was used for quantitative RT-PCR analysis using the primer pairs listed in Supplemental Table 1 and SYBR Green in an ABI7500 machine (Applied Biosystems). Amplification of ACT7 served as a control to estimate relative expression levels.
Genomic DNA Extraction and Bisulfite Sequencing
Genomic DNA was extracted from 3-week-old seedlings (100 mg) using the DNeasy Plant Mini Kit (Qiagen). About 500 to 800 ng of genomic DNA was then used for bisulfite conversion using the EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s instructions. Bisulfite-treated DNA (5 µL) was then amplified by PCR in a 25-μL reaction using ZymoTaq DNA polymerase (ZYMO Research) and degenerate primers (Supplemental Table 1) targeting the −382 to −39 CCA1 promoter region containing the G-box and the CHE binding site (a motif region spanning −280 to −230). Bisulfite sequencing was also performed using a 5′ forward primer targeting the 5′ UTR of CCA1 and a gene-specific 3′ primer targeting either the endogenous CCA1 coding region or the transgenic LUC coding region (Supplemental Figure 3A). PCR products were then resolved on a 1% agarose gel, excised, purified using the UltraClean DNA Purification Kit (MO BIO Laboratories), and cloned into a pGEM-T vector (Promega) for sequencing. For each plant genotype, 14 to 20 independent top-strand clones were sequenced. Bisulfite DNA sequences and the levels of DNA methylation at the CCA1 promoter were analyzed using the online Kismeth program (Gruntman et al., 2008). For each genotype, the percentage of cytosine methylation in each context (CG, CHH, or CHG) was calculated, and the difference in DNA methylation between two genotypes was analyzed using Student’s t test. In Supplemental Figure 2, an integrative genome browser was used to display methylation levels at the single nucleotide level (Robinson et al., 2011). Methylation at the ASA1 locus was used as a control for the bisulfite conversion (∼95%) (Jeddeloh et al., 1998), and the methylation levels of the endogenous ASA1 locus were similar between the Ler × C24 and LerC × C24 reciprocal hybrids (Supplemental Figure 4).
Starch and Biomass Analysis
Starch content was measured in rosette leaves from a pool of five to six plants (∼100 to 300 mg fresh weight) as one biological replication according to a published protocol (Ni et al., 2009) (http://www.nature.com/protocolexchange/protocols/521; doi:10.1038/nprot.2009.12). Three replicates were used in each assay. In each replicate, total starch was quantified using 30 μL of the insoluble carbohydrate fraction using a kit from Boehringer Mannheim (R-Biopharm).
Whole rosettes from hybrids and parents were harvested at ∼3 weeks of age (before bolting) and placed in Lawson #217 hybridization bags (Lawson Bags). The weight from aerial rosette leaves was determined after drying the plants at 80°C for 24 h. Aerial rosettes of 6 to 15 plants in three biological replicates were weighed individually, and the average was used to calculate sd (Miller et al., 2012).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: CCA1 (At2g46830), LHY (At1g01060), TOC1 (At5g61380), CHE (At5g08330), ACT7 (At5g09810), and ASA1 (At1g19920).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. CCA1 Expression, Plant Size, and Bisulfite Sequencing Analysis of DNA Methylation in Reciprocal Hybrids.
Supplemental Figure 2. Bisulfite Sequencing Analysis of DNA Methylation in Reciprocal Hybrids.
Supplemental Figure 3. Bisulfite Sequencing Analysis of DNA Methylation at the 5′ UTR of CCA1.
Supplemental Figure 4. Bisulfite Sequencing Analysis of DNA Methylation at ASA1.
Supplemental Figure 5. Changes in DNA Methylation and Circadian Gene Expression.
Supplemental Figure 6. Biomass Analysis in Reciprocal Hybrids and Their Parents.
Supplemental Figure 7. Analyses of CCA1 Expression and DNA Methylation in Reciprocal Hybrids.
Supplemental Figure 8. Diurnal Expression of Clock Regulators in Developing Siliques in Arabidopsis (Col-0 and Ler).
Supplemental Figure 9. Parent-of-Origin Effects of ProCCA1:LUC Expression in Embryos of Reciprocal Hybrids.
Supplemental Table 1. Primers for Quantitative RT-PCR, PCR with CAPS, and Methylation Assays.
Supplementary Material
Acknowledgments
We thank Eric Richards for providing ddm1-2 and met1-1 mutant seeds and Changqing Zhang for providing a modified plasmid that we used to make the CCA1:LUC construct. This work was supported by the National Science Foundation (Grant IOS1238048 to Z.J.C. and Grant IOS1025965 to C.R.M.).
AUTHOR CONTRIBUTIONS
Z.J.C., M.M., D.W.-K.N., H.H.Y., and C.R.M. designed experiments. D.W.-K.N., M.M., H.H.Y., E.-D.K., T.-Y.H., J.L., and Q.X. performed experiments. D.W.-K.N., M.M., H.H.Y., Q.X., C.R.M., and Z.J.C. analyzed data. Z.J.C., D.W.-K.N., M.M., H.H.Y., and C.R.M. wrote the article.
Glossary
- RdDM
RNA-directed DNA methylation
- siRNA
small interfering RNA
- Ler
Landsberg erecta
- Col-0
Columbia-0
- LerC
Landsberg erecta (CCA1:LUC or ProCCA1:LUC) transgenic line
- ColC
Columbia-0 (CCA1:LUC or ProCCA1:LUC) transgenic line
- C24C
C24 (CCA1:LUC or ProCCA1:LUC) transgenic line
- UTR
untranslated region
- MS
Murashige and Skoog
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adams S., Vinkenoog R., Spielman M., Dickinson H.G., Scott R.J. (2000). Parent-of-origin effects on seed development in Arabidopsis thaliana require DNA methylation. Development 127: 2493–2502. [DOI] [PubMed] [Google Scholar]
- Aufsatz W., Mette M.F., van der Winden J., Matzke A.J., Matzke M. (2002). RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 (suppl. 4): 16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass J., Takahashi J.S. (2010). Circadian integration of metabolism and energetics. Science 330: 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Yao H., Chudalayandi S., Vaiman D., Veitia R.A. (2010). Heterosis. Plant Cell 22: 2105–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Aufsatz W., Zilberman D., Mette M.F., Huang M.S., Matzke M., Jacobsen S.E. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13: 2212–2217. [DOI] [PubMed] [Google Scholar]
- Chen Z.J. (2010). Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 15: 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J. (2013). Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 14: 471–482. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith A.C. (2011). Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12: 565–575. [DOI] [PubMed] [Google Scholar]
- Fujimoto R., Taylor J.M., Shirasawa S., Peacock W.J., Dennis E.S. (2012). Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc. Natl. Acad. Sci. USA 109: 7109–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., et al. (2010). An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature 465: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A., Smith A.M. (2011). Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 16: 169–175. [DOI] [PubMed] [Google Scholar]
- Graf A., Schlereth A., Stitt M., Smith A.M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 107: 9458–9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groszmann M., Greaves I.K., Albertyn Z.I., Scofield G.N., Peacock W.J., Dennis E.S. (2011). Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. USA 108: 2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntman E., Qi Y., Slotkin R.K., Roeder T., Martienssen R.A., Sachidanandam R. (2008). Kismeth: Analyzer of plant methylation states through bisulfite sequencing. BMC Bioinformatics 9: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J.R., Pikaard C.S. (2011). Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 12: 483–492. [DOI] [PubMed] [Google Scholar]
- Haig D. (2013). Kin conflict in seed development: An interdependent but fractious collective. Annu. Rev. Cell Dev. Biol. 29: 189–211. [DOI] [PubMed] [Google Scholar]
- Han Z., Mtango N.R., Patel B.G., Sapienza C., Latham K.E. (2008). Hybrid vigor and transgenerational epigenetic effects on early mouse embryo phenotype. Biol. Reprod. 79: 638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60: 357–377. [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. [DOI] [PubMed] [Google Scholar]
- Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C. (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308: 118–120. [DOI] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- Huh J.H., Bauer M.J., Hsieh T.F., Fischer R.L. (2008). Cellular programming of plant gene imprinting. Cell 132: 735–744. [DOI] [PubMed] [Google Scholar]
- James A.B., Monreal J.A., Nimmo G.A., Kelly C.L., Herzyk P., Jenkins G.I., Nimmo H.G. (2008). The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322: 1832–1835. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Bender J., Richards E.J. (1998). The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 12: 1714–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeddeloh J.A., Stokes T.L., Richards E.J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22: 94–97. [DOI] [PubMed] [Google Scholar]
- Kankel M.W., Ramsey D.E., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J. (2003). Arabidopsis MET1 cytosine methyltransferase mutants. Genetics 163: 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y., Matsui A., Kawashima M., Kaminuma E., Ishida J., Morosawa T., Mochizuki Y., Kobayashi N., Toyoda T., Shinozaki K., Seki M. (2008). Identification of the candidate genes regulated by RNA-directed DNA methylation in Arabidopsis. Biochem. Biophys. Res. Commun. 376: 553–557. [DOI] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Du J., Hale C.J., Feng S., Krajewski K., Palanca A.M., Strahl B.D., Patel D.J., Jacobsen S.E. (2013). Polymerase IV occupancy at RNA-directed DNA methylation sites requires SHH1. Nature 498: 385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J., Steinfath M., Meyer R.C., Selbig J., Melchinger A.E., Willmitzer L., Altmann T. (2009). Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. Plant J. 59: 777–788. [DOI] [PubMed] [Google Scholar]
- Lu J., Zhang C., Baulcombe D.C., Chen Z.J. (2012). Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc. Natl. Acad. Sci. USA 109: 5529–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C.R. (2006). Plant circadian rhythms. Plant Cell 18: 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K., Chandler V., Cone K., Kaeppler H., Kaeppler S., Kerschen A., Pikaard C., Richards E., Sidorenko L., Smith T., Springer N., Wulan T. (2005). Transgene-induced RNA interference as a tool for plant functional genomics. Methods Enzymol. 392: 1–24. [DOI] [PubMed] [Google Scholar]
- Meyer R.C., Törjék O., Becher M., Altmann T. (2004). Heterosis of biomass production in Arabidopsis. Establishment during early development. Plant Physiol. 134: 1813–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R.C., et al. (2012). Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J. 71: 669–683. [DOI] [PubMed] [Google Scholar]
- Miller M., Zhang C., Chen Z.J. (2012). Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 (Bethesda) 2: 505–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T., Haig D. (1991). Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 7: 45–49. [DOI] [PubMed] [Google Scholar]
- Mosher R.A., Melnyk C.W., Kelly K.A., Dunn R.M., Studholme D.J., Baulcombe D.C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286. [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nagel D.H., Kay S.A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22: R648–R657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y., Sahar S., Astarita G., Kaluzova M., Sassone-Corsi P. (2009). Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324: 654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z., Kim E.D., Ha M., Lackey E., Liu J., Zhang Y., Sun Q., Chen Z.J. (2009). Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature 457: 327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordally Z.B., Ishii K., Atkins K.A., Wetherill S.J., Kusakina J., Walton E.J., Kato M., Azuma M., Tanaka K., Hanaoka M., Dodd A.N. (2013). Circadian control of chloroplast transcription by a nuclear-encoded timing signal. Science 339: 1316–1319. [DOI] [PubMed] [Google Scholar]
- Onodera Y., Haag J.R., Ream T., Costa Nunes P., Pontes O., Pikaard C.S. (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120: 613–622. [DOI] [PubMed] [Google Scholar]
- Penfield S., Hall A. (2009). A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21: 1722–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz J.D., Straume M., Stanewsky R., Jamison C.F., Brandes C., Dowse H.B., Hall J.C., Kay S.A. (1997). Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12: 204–217. [DOI] [PubMed] [Google Scholar]
- Prasai M.J., George J.T., Scott E.M. (2008). Molecular clocks, type 2 diabetes and cardiovascular disease. Diab. Vasc. Dis. Res. 5: 89–95. [DOI] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raissig M.T., Baroux C., Grossniklaus U. (2011). Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011). Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., He H., Li J., Chen W., Wang X., Guo L., Peng Z., He G., Zhong S., Qi Y., Terzaghi W., Deng X.W. (2012). Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24: 875–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Fulton D.C., Chia T., Thorneycroft D., Chapple A., Dunstan H., Hylton C., Zeeman S.C., Smith A.M. (2004). Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 136: 2687–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G.S., et al. (2010). Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol. Plant 3: 1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek F.W., et al. (2005). Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger M., Heimes S., Riedel L., Sänger H.L. (1994). RNA-directed de novo methylation of genomic sequences in plants. Cell 76: 567–576. [DOI] [PubMed] [Google Scholar]
- Wijnen H., Young M.W. (2006). Interplay of circadian clocks and metabolic rhythms. Annu. Rev. Genet. 40: 409–448. [DOI] [PubMed] [Google Scholar]
- Xiao W., Brown R.C., Lemmon B.E., Harada J.J., Goldberg R.B., Fischer R.L. (2006). Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol. 142: 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M.Y., Hsieh P.H., Coleman-Derr D., Eshed-Williams L., Thao K., Harmer S.L., Zilberman D. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., Ecker J.R. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao X., Johansen L.K., Xie Z., Carrington J.C., Jacobsen S.E. (2004). Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 14: 1214–1220. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 39: 61–69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.