Abstract
Summary: There have been numerous applications developed for decoding and visualization of ab1 DNA sequencing files for Windows and MAC platforms, yet none exists for the increasingly popular smartphone operating systems. The ability to decode sequencing files cannot easily be carried out using browser accessed Web tools. To overcome this hurdle, we have developed a new native app called DNAApp that can decode and display ab1 sequencing file on Android and iOS. In addition to in-built analysis tools such as reverse complementation, protein translation and searching for specific sequences, we have incorporated convenient functions that would facilitate the harnessing of online Web tools for a full range of analysis. Given the high usage of Android/iOS tablets and smartphones, such bioinformatics apps would raise productivity and facilitate the high demand for analyzing sequencing data in biomedical research.
Availability and implementation: The Android version of DNAApp is available in Google Play Store as ‘DNAApp’, and the iOS version is available in the App Store. More details on the app can be found at www.facebook.com/APDLab; www.bii.a-star.edu.sg/research/trd/apd.php
The DNAApp user guide is available at http://tinyurl.com/DNAAppuser, and a video tutorial is available on Google Play Store and App Store, as well as on the Facebook page.
Contact: samuelg@bii.a-star.edu.sg
1 INTRODUCTION
Single-pass sequencing is a routinely used technique for many areas of biological research. It is used to assess the accuracy of automated sequencing outputs and for further analyses such as primer designs, alignments and BLAST. To meet with the demands of sequencing services that generate these files (Applied Biosystems or ab1 sequencing files), many software tools (both freely and commercially available) have been created in the past decade for Windows/MAC/Linux Operating Systems (OS). Some examples of such software include BioEdit (Hall, 1999, 2011; see www.mbio.ncsu.edu), Chromas (http://www.technelysium.com.au/), FinchTV (http://www.geospiza.com) and SeqAssem (www.sequentix.de). Given the increasing popularity of (iOS and Android) smartphone and tablet users in recent years see (https://smallbusiness.yahoo.com/advisor/smartphone-tablet-continues-rise-stats-210509525.html), Jaume, 2013; it is interesting that such bioinformatics tools have yet to thoroughly penetrate such OS and devices. With the recent rise of iPhone/iPad and Android devices, scientists, along with others, have been benefiting from the convenience of mobility in their everyday lives (Comstock, 2014; see http://mobihealthnews.com/29253/survey-doctors-prefer-tablets-for-journal-articles-smartphones-for-most-other-tasks/), Dufau et al., 2011; Evanko, 2010; Sutton and Fraser, 2013; see http://lifescientist.com.au/content/biotechnology/article/the-rise-of-smartphone-health-and-medical-apps-1072193834). To raise productivity and expedite research, we have started to develop bioinformatics applications for these two OS. Keeping in mind that the purpose of smartphone apps is for convenience, we have developed a stripped-down version of software such as Bioedit and Chromas (we offer protein translation in addition to other features offered by ChromasLite). DNAApp is the first Android native app to assist in the manipulation and analysis of sequencing data.
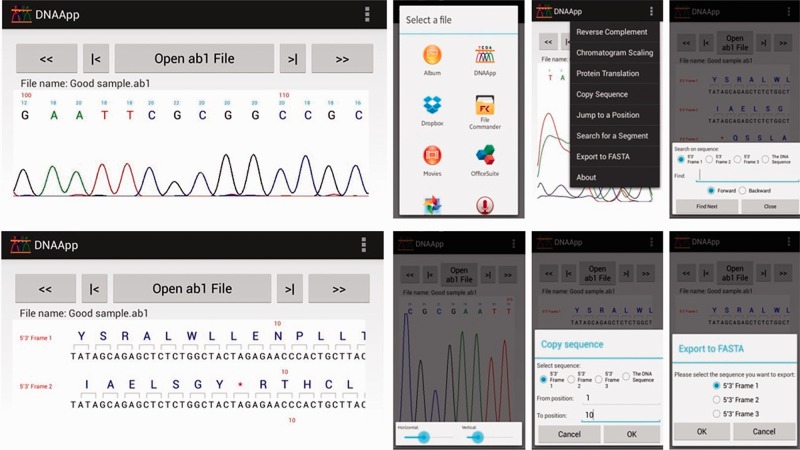
DNAApp runs on both Android and iOS, enabling the decoding of ab1 sequencing files to display the DNA sequence and the corresponding chromatogram. This facilitates a broader range of analysis when used with Web tools such as BLAST and ClustalW. To use it, the user simply has to specify an ab1 file for analysis from any location in the phone memory or from cloud storage like Dropbox. To accommodate the different signal strengths generated for each single-pass sequencing reaction, touch screen functions for both vertical and horizontal scaling of the chromatograms are available. Should the user face difficulties in the touch screen scaling, a scaling slider was also built in to facilitate chromatogram scaling. This is useful, as the two ends of the sequencing files may have decreased signals, resulting in misreads of nucleotide bases. In addition, to facilitate the analysis of sequence quality, the quality score (in percentage scale) of each base was also included. For the analysis of sequencing data generated by reverse primers, we incorporated a reverse complement function along with its inferred chromatogram. Should the user desire further analysis using browser-based bioinformatics tools (such as Expasy, BLAST, ClustalW), the ‘user-defined cut and paste’ function would come in handy, overcoming the highly difficult task of highlighting sequences in a touch-screen interface. Similarly, for jumping to specific locations of the long data output (typically, at least 700 bases), we have a user-defined ‘jump to’ function, as well as a search function for sequence segments of interest [including primers, restriction sites and tags; see Fig. 1 for the Android graphical user interface (GUI) of DNAApp and supporting features]. To aid users in further analysis, DNAApp is capable of generating all six possible translations of a DNA sequence to proteins. The copy and search functions are applicable for protein sequences. Should the user require storing the sequence data as a human-readable text-based document, the DNAApp allows the export of FASTA format files. Because the analyzed sequence would be too long to fit within the device screen in a single line, we incorporated the use of fast and end scrolling icons for different speed scrolling. For longer reads, the app also adapts by autorotation to landscape orientation.
Fig. 1.
The friendly GUI of DNAApp and its supporting features on Android
In the development of DNAApp, the biggest challenge was the decoding of the sequencing ab1 file. Using the ABIF (Applied Biosystem Inc. Format), the elements of data stored in the ab1 file include the trace data of the four signal channels corresponding to DNA bases adenine (A), guanine (G), thymine (T) and cytosine (C); the array of peak locations; the DNA sequence inferred by the base caller of the sequencing system; and the quality score array. DNAApp acquires each data item by looking up the desired entry in the file directory, figures out the starting offset, data size of each data element and number of elements the data item has and finally performs the reading. Chromatograms are shown by plotting the arrays of trace data of the four channels A, G, T and C using spline interpolation (Ahlberg and Nilson, 1967). To generate the reverse complement chromatogram, inference was done by:
, in which , where is the complement base of b, L is the sequence length, 0 ≤ i ≤ L-1. This feature assesses the confidence of reverse complement bases. For the segment searching feature, we used the Knuth-Morris-Pratt algorithm (Knuth et al., 1977)
DNAApp is created for ‘on-the-go’ visualization and quick analysis of ab1 files on mobile computing platforms. It has been built to facilitate integration with online Web tools (accessible through mobile browsers) for in-depth analysis if needed. Examples of such analyses that can be performed are available in the app download pages and user guide.
The Android version of the application is available in Google Play Store for Android version 4.4.2 onward and is compatible with both smart phones and tablet PCs at configurations as low as 1Ghz CPU and 512MB RAM. The iOS version is available in App Store for iOS 7 and above.
FUTURE DEVELOPMENT
Additional applications will be developed with related features in the near future for the next generation of bioinformatics tools. More information can be obtained from www.facebook.com/APDLab; www.bii.a-star.edu.sg/research/trd/apd.php
ACKNOWLEDGEMENTS
The authors thank Mr Keane Lim for the writing of the DNAApp user guide.
Funding: This work is funded by the Joint Council Office, Agency for Science, Technology, and Research, Singapore
Conflict of interest: none declared.
REFERENCES
- Ahlberg J, Nilson E. The theory of splines and their applications. New York, USA: Elsevier Academic Press; 1967. [Google Scholar]
- Comstock J. Survey: Doctors Prefer Tablets For Journal Articles, Smartphones For Most Other Tasks. Mobi Health News: 2014. . http://mobihealthnews.com/29253/survey-doctors-prefer-tablets-for-journal-articles-smartphones-for-most-other-tasks/ (18 August 2014, date last accessed) [Google Scholar]
- Dufau S, et al. Smart phone, smart science: how the use of smartphones can revolutionize research in cognitive science. PloS One. 2011;6:e24974. doi: 10.1371/journal.pone.0024974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanko D. The scientist and the smartphone. Nat. Methods. 2010;7:87. [Google Scholar]
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hall T. BioEdit: an important software for molecular biology. GERF Bull. Biosci. 2011;2:60–61. [Google Scholar]
- Jaume J. Smartphone and tablet use continues to rise: the stats. Yahoo Small Business Advisor. 2013 [Google Scholar]
- Knuth D, et al. Fast pattern matching in strings. SIAM J. Comput. 1977;6:323–350. [Google Scholar]
- Sutton M, Fraser M. The rise of smartphone health and medical apps. Australian Life Scientist. 2013 [Google Scholar]