Abstract
Purpose
We propose a systematic approach to correlate MRI and digital histopathology in prostate.
Methods
T2-weighted (T2W) MRI and diffusion weighted imaging (DWI) are acquired, and a patient-specific mold (PSM) is designed from the MRI. Following prostatectomy, a whole mount tissue specimen is placed in the PSM and sectioned, ensuring tissue blocks roughly correspond to MRI slices. Rigid-body and thin plate spline (TPS) deformable registration attempt to correct deformation during image acquisition and tissue preparation and achieve a more complete one-to-one correspondence between MRIs and tissue sections. Each tissue section is stained with hematoxylin and eosin and segmented by adopting a machine-learning approach. Utilizing this tissue segmentation and image registration, the density of cellular and tissue components (lumen, nucleus, epithelium, and stroma) is estimated per MR voxel, generating density maps for the whole prostate.
Results
This study was approved by the local IRB and informed consent was obtained from all patients. Registration of tissue specimens and MRIs was aided by the PSM and subsequent image registration. Tissue segmentation was performed using a machine-learning approach, achieving ≥0.98 AUCs for lumen, nucleus, epithelium and stroma. Examining the density map of tissue components, significant differences were observed among cancer, benign peripheral zone, and benign prostatic hyperplasia (BPH) (p-value<5e-2). Similarly, the signal intensity of the corresponding areas in both T2W MRI and DWI was significantly different (p-value<1e–10).
Conclusions
The proposed approach is able to correlate MRI and digital histopathology of the prostate and is promising as a potential tool to facilitate a more cellular and zonal tissue-based analysis of prostate MRI, based upon a correlative histopathology perspective.
Keywords: prostate, histopathology, image registration, machine learning
1. INTRODUCTION
Prostate cancer is one of the most prevalent cancers in US men, and the second leading cause of cancer death [1]. Widespread screening has increased the incidence of prostate cancer, and qualitative histological assessment is the basis of cancer diagnosis today. Like all qualitative measurements, histologic interpretation is subject to interobserver variation and is difficult to measure for purposes of research [2] [3]. Several imaging techniques such as magnetic resonance imaging (MRI) and positron emission tomography (PET) provide minimally-invasive means to evaluate and characterize prostatic tissue. MRI, in particular, offers a (relatively) high spatial resolution and soft tissue contrast. The multi-parametric MRI, including T2-weighted (T2W) MRI, diffusion weighted MRI (DWI), dynamic contrast-enhanced (DCE) MRI, and MR spectroscopy, is capable of identifying cancers with high sensitivity of 80–95% or more depending on the criteria used [4] [5]. In prostate pathology, the microscopic tissue properties are of great importance; however, detailed tissue or cellular information is not feasible in MRI due to its resolution of ~300μm. Digital histopathology offers the ability to quantitatively measure tissue characteristics with the resolution of ~0.5μm (at 20× magnification); for instance, distortion of glandular structure and spatial distribution and arrangement of cells and nuclei. Such quantitative measures can be efficiently and effectively integrated into a decision-support system for accurate diagnosis and prognosis utilizing machine-learning techniques. Previous studies have suggested that the digital pathology-based image features could improve cancer diagnosis and prognosis [6] [7] [8]. Accordingly, a combination of a multi-parametric MRI approach and digital histopathology analysis could facilitate characterization of prostate cancer and an improved understanding of the underlying origins of MRI features. The approach necessitates an accurate registration between the two techniques, but it is often limited due to the substantial difference in the resolution between them.
Several studies have been conducted to examine the relationship between MRI and prostate histopathology. T2W MRI was investigated in relation to tissue density [9] and to histopathology (i.e. carcinoma and benign prostatic hyperplasia (BPH)) [10]. Apparent diffusion coefficients (ADC) derived from DWI has been shown to be inversely related to nuclear density [11] [12] [13] [14]. Different tissue components (e.g., cytoplasm, stroma, lumen, and nuclei) were also correlated with T2W MRI, DWI, and DCE MRI [14]. Moreover, Gleason score was inversely related to ADC [15] and T2 measurements [13]. For these studies, the regions of interest (ROIs) were manually selected and compared by pathologists and radiologists using consensus. Such manual region-to-region analysis may potentially introduce bias to the analysis and thus, limit its value.
Herein, we propose a systematic approach to correlate a digital histopathology analysis with a multi-parametric MRI (Figure 1). We adopt a machine learning approach to conduct digital histopathologic analysis of the prostate. The corresponding MR slices and tissue specimen images are registered via a patient-specific mold (PSM) and semi-automated deformable image registration. A complete density map for different tissue components is generated for the whole prostate in MRI.
Figure 1.
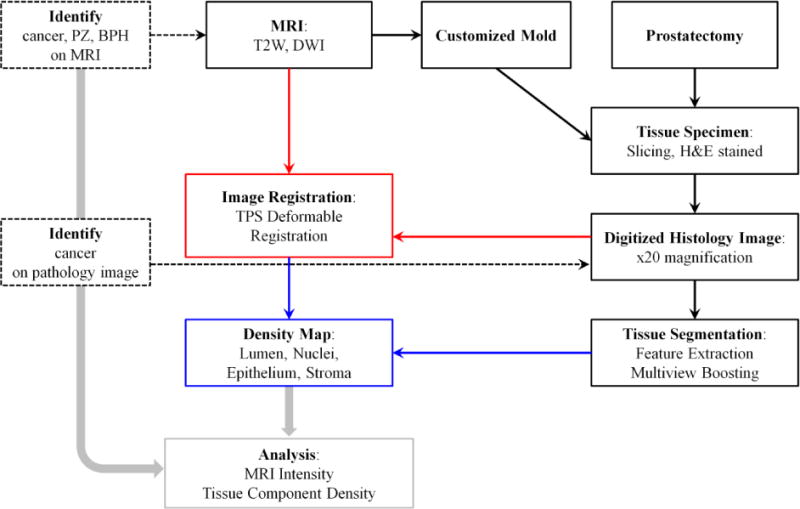
Block diagram for the method of correlating MRI with digital histopathology.
2. METHODS
2.1. Patient Population
This study was approved by the local institutional review board (IRB) and was compliant with the Health Insurance Portability and Accountability Act. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2008 (5). We included 8 consecutive radical prostatectomy patients with 15 tissue sections (obtained between March 2014 to July 2014), where pre-surgical MR images are free from substantial artifacts, distortion, noise, or partial volume effects, in this study. All patients had biopsy proven adenocarcinoma of prostate and underwent robotic assisted radical prostatectomy within 180 days of imaging without any intervening treatment. Informed consent was obtained from all patients for being included in the study.
2.2. MRI Protocol
Multi-parametric MRI of the prostate is performed on a 3-Tesla MR scanner (Achieva-TX, Philips Healthcare, Best, NL) using the anterior half of a 32-channel SENSE cardiac coil (In Vivo, Philips Healthcare, Gainesville FL, USA) and an endorectal coil (BPX-30, Medrad, Indianola PA, USA). No pre-examination bowel preparation was required. The balloon of each endorectal coil is distended with approximately 45 mL of perfluorocarbon (Fluorinert FC-770, 3M, St Paul, MN, USA) to reduce imaging artifacts related to air-induced susceptibility. T2-weighted (T2W) MRI and diffusion-weighted MRI (DWI) are acquired. T2W MRI has a resolution of 0.27mm × 0.27mm. The standard DWI is acquired with 5 evenly-spaced b-values (0–750 s/mm2), and a map of the apparent diffusion coefficient (ADC) is calculated per voxel. Multi-parametric MRI is independently evaluated by three experienced genitourinary radiologists (SS, BT, and PLC with 2, 7 and 14 years of experience). The whole prostate, peripheral zone, transition zone, and cancer lesions are delineated and recorded in an MRI coordinate system. The whole prostate is first automatedly segmented by research software (iCAD Inc., Nashua, NH, USA) and the resulting segmentation is manually adjusted by the radiologists. DWI images are rigidly registered with T2W MRI images using MR coordinate information [16]. The registration is performed per MR slice.
2.3. Customized MRI based Mold and Tissue Specimen Preparation
The patient-specific mold (PSM) is printed based on the pre-surgical MRI of each patient, ensuring that the tissue blocks correspond to the MRI slices [17]. The PSM is created in SolidWorks, a 3D-CAD software (Dassault Systems SolidWorks Corp., Walthan, MA, USA) and printed on a 3D-printer. The PSM is designed with sectioning slots that are positioned such that each tissue block matched the location of a 3mm thick MR slice.
Following prostatectomy, a whole mount prostate tissue specimen is fixed in formalin for 2–24h at room temperature. The tissue specimen is serially sectioned in the PSM from apex to base at 6mm intervals using a 10 inch long autopsy knife (Scientific Supplies, order No. MII-5000). The resulting tissue blocks are labeled and allowed to fix in formalin for an additional 48–72 h. The formalin is removed by wax addition and graded alcohol is added to dehydrate the specimen. The alcohol is cleared by adding xylene and paraffin to the media. Tissue sections are cut at 5μm thickness and stained with hematoxylin-eosin (H&E) for histopathologic evaluation. A digital image of each tissue section is acquired on a standard optical microscope (Aperio Technologies, Inc.) at 20× magnification. The image has a resolution of 0.504μm × 0.504μm and an approximate dimension of 100,000 × 80,000 pixels (or 5cm × 4cm).
On the 18 digitized tissue images from 4 patients, lumen, nuclei, epithelium, and stroma pixels are manually selected by pathologists and used for training and testing tissue segmentation methods. The training dataset contains 23,359 lumen, 27,570 nuclei, 80,809 epithelium, and 70,545 stroma pixels from 2 patients’ 11 tissue images. The validation dataset consists of 7 tissue images from 2 patients, including 16,482 lumen, 19,342 nuclei, 57,289 epithelium, and 46,991 stroma pixels.
2.4. Tissue Image Analysis
2.4.1. Feature Extraction
A tissue specimen image I (in RGB: red, green, and blue) is converted into 3 different forms: 1) Histogram equalization 2) HSV (hue, saturation, and value) color space 3) La*b* (L: illumination, a* and b*: color-opponent dimensions) color space. In total, 9 color channels are generated. For a pixel x∈I, we compute intensity- and texture-based features within a w × w rectangular window around the pixel x. Intensity-based features include average, standard deviation, kurtosis, and skewness. Texture-based features utilize local binary pattern (LBP) [18], local directional derivative pattern (LDDP) [19], and variance measure (VAR) [18], that generate a (binary) pattern code per pixel. The pattern codes in a window are summarized into a histogram and used as texture-based features. These features are computed on the two color channels (HSV value and La*b* illumination) using two neighborhood topologies (P,R)={(16,2),(24,3)} where P and R are the number of neighboring pixels and the radius, respectively. Concatenating the intensity- and texture-based features, we obtain a K-dimensional feature vector f (x) = {fk (x) |k ∈ {1,2, …,K}}, where fk (x) is the value of feature k at pixel x∈I. We perform the feature extraction for differing sizes of w × w window (w=1, 3, 7, 15, 27, 43, and 63 pixels) and obtain a set of feature vectors , where fm is a feature vector in a view m; f1 constitutes 9 features that are intensity values of 9 color channels (w=1). f2 includes 36 features (average, standard deviation, kurtosis, and skewness per color channel; w=3). For m≥3, fm generates 252 features that consist of 36 intensity-based features and 216 texture-based features ((P,R)=(16,2): 18 features for LBP and LDDP and 10 features for VAR per color channel; (P,R)=(24,3): 26 features for LBP and LDDP and 10 features for VAR per color channel).
2.4.2. Local Binary Pattern and Its Variants
For a (center) pixel x∈I, LBP examines its neighboring pixels p (p = 0, …, P − 1) in a radius R and generates a binary pattern code as follows:
| (1) |
where s (x) is 1 if x ≥ 0 and 0 if x < 0 and gc and gp represent the gray level of the center pixel and its neighborhood pixels, respectively. The coordinates of the neighborhood pixels are computed as (R cos(2πp/P), − R sin (2πp/P)) and their gray levels are estimated by interpolation. Since LBP exclusively depends on the sign of the gray level differences, the pattern code is invariant to the scaling of the gray scale. Moreover, rotating the image, the gray level of the neighborhood pixels rotate around the center pixel. The rotation results in a different binary pattern code but only makes a bit-wise shift in the original pattern code. Hence, rotation-invariant pattern code is computed as
| (2) |
where ROR(x, i) is a circular bit-wise shift operator on x by i bits.
Higher order derivative information is computed using local directional derivative pattern (LDDP) to provide more detailed texture information. 2nd order LDDP along p direction is computed as follows:
| (3) |
| (4) |
where and denote the gray level of a neighborhood pixel p in a circle of radius R1 and R2, respectively.
Due to lack of contrast information in LBP and LDDP, variance of the local contrast (VAR) is also measured as follows:
| (5) |
Since VAR is continuous, it is discretized by equal-depth binning to provide a pattern code (We set 10 bins).
2.4.3. Multiview Boosting
Multiview boosting algorithm [20] takes a dataset of differing views and cooperatively integrates them together to construct a (strong) classifier in a boosting scheme. It maintains a cost matrix (or distribution) C of data for each view and a global cost matrix CG in a way that the harder cases for one view are managed by the other views. At each booting round, it learns a weak classifier on each view and selects the best one. At the end of the boosting iterations, a weighted vote of the chosen weak classifiers forms the final classifier.
Consider a dataset S = {(x1, y2), …, (xn, yn)} where xi ∈ I is a pixel and yi ∈ Y is a histology class label. A pixel xi in a view m, designated as xi,m, generates a feature vector fi,m. C is a cost matrix where C (i,l) is the cost of classifying xi as the class l. If a prediction is correct, the cost gets lower, otherwise higher. One cost matrix Cj per view j and a global cost matrix CG are defined. Multiview boosting runs for T iterations as follows (T=20).
First, a weak classifier ht,j is learned on a cost matrix Ct,j (or view j) at iteration t, which performs better than an edge-over-random baseline B. That is, ht,j satisfies the edge condition if where 1h is the prediction matrix defined as 1h (i,l) is 1 if h(i)= l and 0 otherwise. B is a cost matrix Uγ defined as Uγ (i,l) is if yi =l and if yi ≠ l.
Second, cost matrices are updated using the weak classifiers. Ct,j(i, l) is updated only if ht,j correctly classifiers xi as the class l or all the other weak classifiers misclassify it:
| (6) |
where ,and
Third, on the global cost matrix Ct,G, the best performing weak classifier ht is chosen and its weight α1 is computed as where the edge δt = Ct,G·Uγ,G − Ct,G·1t,G. Fourth, using the selected weak classifiers so far, Ct,G is updated. Lastly, we obtain the final hypothesis as a weighted vote of the selected weak classifiers: H (x) = argmaxl∈Y QT (x,l) where . As a weak classifier, a linear support vector machine (SVM) [21] is adopted. We compute the probability estimate of the output QT(x) using a logistic link function: . The detail of multiview boosting algorithm is available in [20].
2.4.4. Tissue Segmentation
A tissue specimen image is segmented into lumen, nucleus, epithelium, and stroma in a cascaded fashion. The tissue image is first segmented into lumen and non-lumen areas. Lumens are determined by using a threshold value thL (>0.5) on the output of the lumen vs. non-lumen multiview boosting classification (+: lumen, −: non-lumen), followed by a size constraint sL (>50um2). Second, non-lumen areas are classified into nuclei and non-nuclei areas. Thresholding (thN: >0.5) the output of the nuclei vs. non-nuclei multiview boosting classification (+: nuclei, −: non-nuclei), initial nuclei are identified. The size and shape of the initial nuclei are examined: If the size of a nucleus is smaller than 5μm2 or the ratio of the major and minor axis is greater than 5 when its size is smaller than 25μm2, then the nucleus is considered to be an artifact. Third, non-nuclei areas are grouped into epithelium and stroma. We identify epithelium by using a threshold value thES on the output of the epithelium vs. stroma multiview boosting classification (+: epithelium, −: stroma), followed by a size constraint sES. To reflect and correct the variability in staining and improve the segmentation, we adjust the threshold value and size constraint (thE:0.4–0.6; sE: 500–1500um2) per slice. Fourth, nuclei that are present in epithelium are designated as epithelial nuclei. Finally, the perimeter of lumens is examined. By definition, epithelial cells enclose lumens in tissue. If <40% of the perimeter is surrounded by epithelium, such lumens are excluded. The tissue segmentation was performed in MATLAB on the high-performance Biowulf Linux cluster at the National Institutes of Health (Bethesda, MD, USA) (http:/biowulf.nih.gov).
2.5. Image Registration and Density Map
Although the PSM helps in orienting the tissue specimen and maintaining the shape of the prostate, tissue preparation process (e.g., fixation) introduces deformation. The endorectal coil also deforms the prostate during the MRI and this deformation is absent in the specimen. To correct such deformations, each pair of the MRI slice and whole mount tissue slide is examined and their correspondence confirmed by radiologists. Then, in-house developed semi-automated registration, implemented in NIH OncoNav software, completes the registration based on the outer shape and/or internal structures of the prostate. Given the prostate segmentation contours of both tissue specimen image and MRI image, manual rigid-body registration with 6 degrees of freedom is carried out to match the oblique plane of the 3 dimensional MRI image with the tissue specimen image. 30 landmarks are sampled from each of the segmentation contours, paired, and used to conduct 2 dimensional thin plate spline (TPS) deformable registration [22]. The landmarks are obtained by calculating the intersections between the prostate contour and 30 evenly angled rays (an interval of 12 degrees) which start from the center of the prostate segmentation contour. In addition, the internal anatomical landmarks such as urethra, ejaculatory ducts, and BPH nodules can be added to improve the quality of registration. Smoothing is incorporated in the deformable registration to prevent over fitting. The correspondence between MRI image and tissue specimen image is visually confirmed.
A voxel in a MRI slice approximately corresponds to 540 × 540 pixels in a tissue specimen image. We estimate the density, i.e., percentage area of tissue components (lumen, nuclei, epithelium, and stroma) per MR voxel by computing the corresponding location in a tissue image, drawing a 540 × 540 window around the location, and calculating the ratio of the size of the segmented area to the size of the window. In order to compare MR signal intensities and tissue component densities, cancer, peripheral zone (PZ), and BPH are identified by radiologists and transferred to the corresponding tissue specimen image using the image registration transformation. Also, pathologists independently examine tissue specimen images and identify cancers. The identified regions are further adjusted to avoid any distortion, artifacts, or tears in the tissue specimen. Cancer regions are only included when they are identified by both radiologists and pathologists.
2.6. Statistical Analysis
Data analysis is performed using R software version 2.15.2 (GNU General Public License). The performance of tissue segmentation is measured and evaluated using a receiver operating characteristic (ROC) plot (sensitivity vs. 1-specificity) and the area under the ROC curve (AUC). Bootstrap re-sampling with 2000 repetitions is adopted to assess 95% confidence interval (CI) of AUCs [23]. Statistical significance of MR signal intensity and tissue component density in discriminating different histopathologic areas is determined by Wilcoxon rank-sum test.
3. RESULTS
To segment whole mount prostate tissue specimen images, three multiview boosting classifiers were employed 1) Lumen vs. Non-lumen 2) Nucleus vs. Non-nucleus 3) Epithelium vs. Stroma. Each multiview boosting classifier was trained on the training dataset and tested on the validation dataset. The segmentation result was summarized into a ROC plot, and the AUC and confidence intervals (CI) were computed. We obtained an AUC of 0.99 AUC (95% CI: 0.99–1), 0.98 (95% CI: 0.98–0.98), and 0.98 (95% CI: 0.98–0.98) for Lumen vs. Non-lumen, Nucleus vs. Non-nucleus, and Epithelium vs. Stroma, respectively. The cascaded segmentation was conducted for the whole tissue specimen image (Figure 2). H&E stained image and color-coded segmentation of different histopathologic areas (cancer, peripheral zone, BPH) are presented in Figure 2b.
Figure 2.
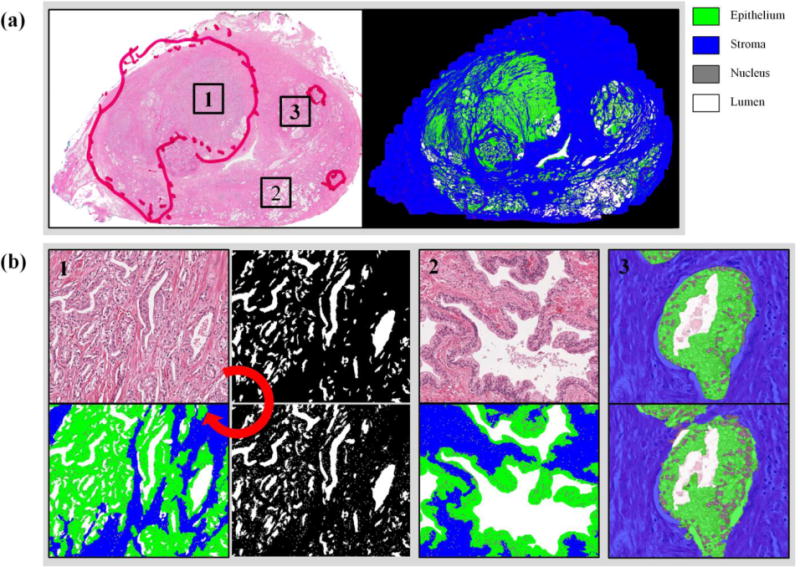
Tissue segmentation. (a) A whole mount prostate tissue specimen is sectioned and stained with hematoxylin and eosin. Color-coded segmentation images are presented. (b) A tissue image is segmented in a cascaded manner: lumen, nuclei, and epithelium and stroma. Tissue images and segmentation results are shown for 1) TZ cancer and 2) benign peripheral zone. 3) The manual segmentation by a pathologist (top) is compared to the automated segmentation result (bottom).
Tissue specimen images were successfully registered with T2W MRI using TPS deformable registration. Since T2W MRI and the ADC map of DWI were already aligned using MR coordinate information, a correspondence between a tissue specimen image and T2W MRI and the ADC map was achieved (Figure 3). Tissue specimen images and MRIs corresponded to each other in tumor lesions and internal structures such as BPH nodules, urethra, ejaculatory ducts, transition zone, and peripheral zone. Moreover, using the deformable registration and tissue segmentation, a density map of the four tissue components (lumen, nuclei, epithelium, and stroma) were computed for the whole prostate (Figure 4).
Figure 3.
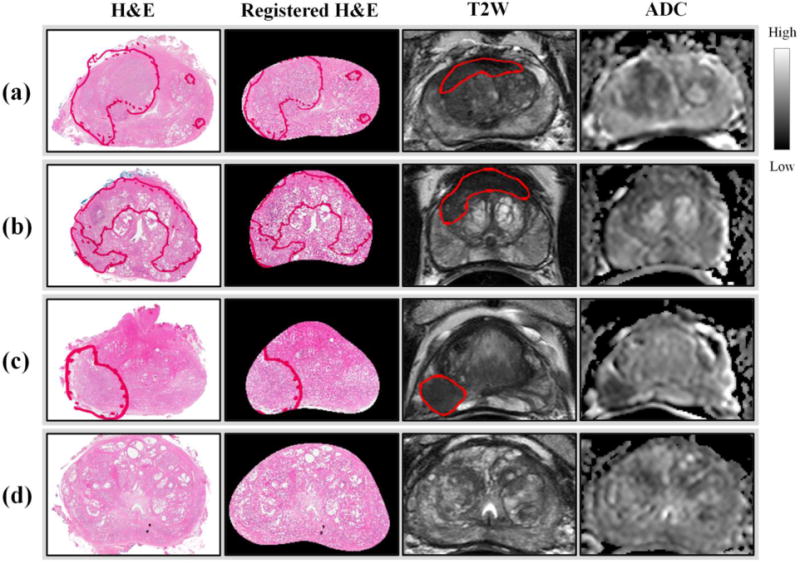
Tissue specimen and MRI registration for 4 different patients. A H&E stained tissue specimen images is registered with its corresponding slice of T2W MRI and the ADC map of DWI. Tumor lesions are marked in red. (a)(b) TZ cancer (c) PZ cancer (d) benign.
Figure 4.
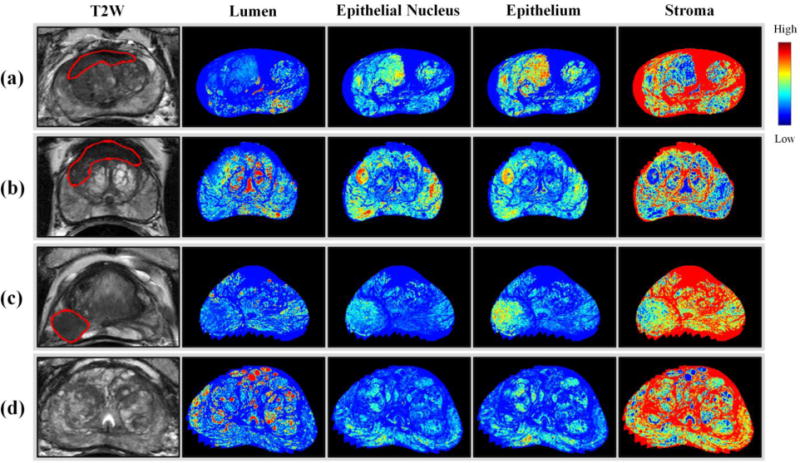
Density maps for 4 different patients. A density map of tissue components is generated for the whole prostate using tissue segmentation and deformable image registration. Tumor lesions are marked in red. (a)(b) TZ cancer (c) PZ cancer (d) benign.
We identified 28 regions (8 transition zone (TZ) cancer, 7 peripheral zone (PZ) cancer, 5 BPH, 8 PZ benign) from 15 tissue sections. 8 cancer regions were from TZ (Gleason score 7(3+4): 3, 8(4+4): 4, 9(4+5): 1), and 7 cancer regions were from PZ (Gleason score 7(3+4): 4, 8(4+4): 3). The MR signal intensities and tissue component densities were compared among four different histopathologic areas (TZ cancer, PZ cancer, BPH, PZ benign). As shown in Figure 5a, for both T2W MRI and the ADC map, the signal intensity in PZ benign was significantly higher than in TZ cancer, PZ cancer, and BPH (p-value<1e-10). A significantly lower signal intensity of the ADC map was obtained in both TZ cancer and PZ cancer than in BPH (p-value<1e-10). PZ cancer showed a higher signal intensity than TZ cancer in the ADC map. Moreover, there were significant differences in density between the histopathologic areas regardless of tissue components (lumen, nucleus, epithelium, and stroma; p-value<5e-2) (Figure 5b). TZ cancer and PZ cancer showed the lower percentage area of lumen and higher percentage area of epithelium and epithelial nuclei. PZ benign had the higher percentage area of lumen and lower percentage area of epithelium and epithelial nuclei. In BPH, the percentage area of lumen and stroma was higher than in both TZ cancer and PZ cancer and lower than in PZ benign; it was lower than both TZ cancer and PZ cancer and higher than PZ benign with respect to the percentage area of epithelium and epithelial nucleus. TZ cancer showed the higher percentage area of epithelium and epithelial nuclei than PZ cancer. The percentage area of stroma had a higher interquartile range (IQR) in the four histopathologic areas as compared to other areas (lumen, epithelium, and epithelial nucleus).
Figure 5.
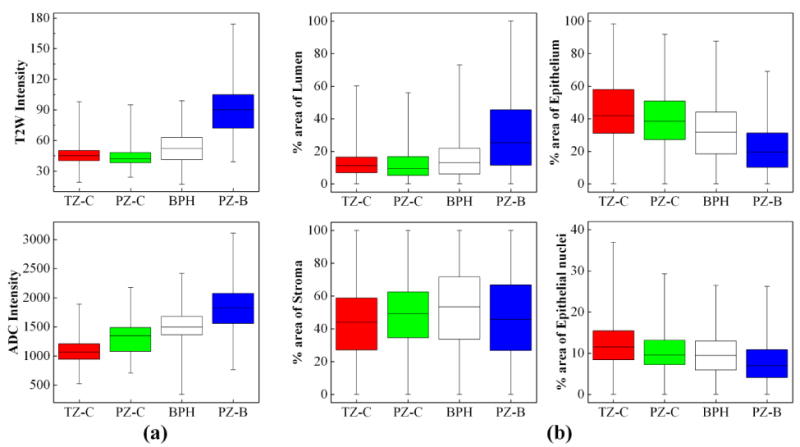
Box plot of MR signal intensity and tissue density. For transition zone cancer (TZ-C), peripheral zone cancer (PZ-C), peripheral zone benign (PZ-B), and benign prostatic hyperplasia (BPH), (a) the signal intensity of T2W and ADC map and (b) the percentage area of different tissue types are plotted.
4. DISCUSSION AND CONCLUSION
Digital histopathology analysis was correlated with multi-parametric MRI of the prostate. Tissue specimen sections were prepared using the PSM to match MRI slices. A multiview boosting classification achieved accurate tissue segmentation in a cascaded fashion. A density map of the four tissue components was computed for the whole prostate through semi-automated deformable registration. This enabled a more complete voxel-to-voxel correspondence between MRI images and tissue specimen images. We observed different characteristics of histopathologic areas in terms of MR signal intensity and tissue component density.
In the previous studies, MRI signal intensities and tissue component densities were examined. The previous findings are consistent with our results. Cancer tissue had the lower signal intensity of the ADC map than benign tissue [11] [12] [13] [14]. The signal intensity of the ADC map in BPH was higher than in cancer tissue and lower than in benign tissue [11]. The signal intensity of T2W MRI in cancer tissue was lower than in benign tissue. The cellular density was higher in cancer tissue than in benign tissue [12] [13] [14]. Cancer tissue showed the lower lumen density than benign tissue [14].
The mainstay of MRI and histopathology correlation studies has been to conduct region-to-region comparisons. The corresponding ROIs were selected on each imaging method. The signal intensity and tissue density were averaged in a region and compared to each other. The ROIs were relatively large and the averaging effect may have suppressed the heterogeneity within each histopathologic area. However, our approach is not limited to such restrictions due to the voxel-to-voxel correspondence. This will aid in characterizing the prostate in MRI in relation to finer histopathologic changes. Local affine transformation-based image registration between tissue specimen images and MRI images was previously presented [24]. It employed quarter mount tissue sections which need manual assembly, and no histopathology analysis was reported.
There are several limitations to this study. First, the sample size is limited. Although roughly one-to-one correlation is available per slice, the prostate is a zonal structure and prostate cancer is a multifactorial, heterogeneous disease with a mixture of heterogeneous growth patterns. The relationship between MRI and tissue component density in different histopathological classifications (e.g., Gleason grades) should be further studied in larger cohorts. Second, although we excluded MR images that possess substantial artifacts, distortion, and noise, these may still exist, leading to inaccurate registration and analyses. Signal inhomogeneity in MRI images may hamper the fair comparison between MRI slices and patients. Additional post-processing on MRI (e.g., normalization) or quantitative maps (e.g., quantitative T2map) can be applied to alleviate their effects and to improve the quality of images and correlation analysis. Third, the image registration between MRI and tissue specimen images relies on the outer shape and anatomical features of the prostate. However, the fragmented tissues or artifacts due to surgery or tissue preparation often degrade the tissue specimen image. These artifacts affect not only the quality of the registration but also the correlation between MR intensity and tissue component density. Fourth, the quality of the image registration was visually evaluated due to the lack of ground truth. Anatomical landmarks (urethra, ejaculatory ducts, and BPH nodules) may be used to measure the accuracy of the registration, but the exact corresponding locations are not available. We rather used them to guide the registration. More robust and accurate methods should be further studied to improve the registration. Fifth, the substantial difference in the resolution between MRI and tissue specimen images is another key challenge to achieve the accurate registration. Such technical constraints may set the upper limit for the registration. But, this was not considered in conducting and evaluating the image registration in this study. Future studies should be conducted in regard to the current technical limitations. Last, only two MRI modalities – T2W MRI and DWI – were considered in this study. Other imaging modalities such as DCE MRI or quantitative T2 mapping can be incorporated in the future using either MR coordinate information or other image registration algorithms.
In conclusion, an approach to correlate whole mount tissue specimens with multi-parametric MRIs is demonstrated. The presented framework could play a critical role in further extending our knowledge of prostate histopathology as expressed by multi-parametric MRI, potentially leading to improved decision-support for MRI and improved understanding of the origins of signal characteristics on MRI. In the future this method could be useful in selecting cells for genomic analysis to further the correlation between MRI and tumor genomics.
Footnotes
CONFLICT OF INTEREST
Peter L. Choyke, Peter A. Pinto, and Bradford J. Wood have a cooperative research and development agreement with Philips Healthcare. Jin Tae Kwak, Sandeep Sankineni, Baris Turkbey, Sheng Xu, and Maria Merino declare that they have no conflict of interest.
References
- 1.Siegel R, Ma JM, Zou ZH, Jemal A. Cancer Statistics, 2014. Ca-Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Glaessgen A, Hamberg H, Pihl CG, Sundelin B, Nilsson B, Egevad L. Interobserver reproducibility of modified Gleason score in radical prostatectomy specimens. Virchows Arch. 2004;445(1):17–21. doi: 10.1007/s00428-004-1034-0. [DOI] [PubMed] [Google Scholar]
- 3.Allsbrook WC, Mangold KA, Johnson MH, Lane RB, Lane CG, Amin MB, Bostwick DG, Humphrey PA, Jones EC, Reuter VE, Sakr W, Sesterhenn IA, Troncoso P, Wheeler TM, Epstein JI. Interobserver reproducibility of Gleason grading of prostatic carcinoma: Urologic pathologists. Hum Pathol. 2001;32(1):74–80. doi: 10.1053/hupa.2001.21134. [DOI] [PubMed] [Google Scholar]
- 4.Turkbey B, Mani H, Shah V, Rastinehad AR, Bernardo M, Pohida T, Pang YX, Daar D, Benjamin C, McKinney YL, Trivedi H, Chua C, Bratslavsky G, Shih JH, Linehan WM, Merino MJ, Choyke PL, Pinto PA. Multiparametric 3T Prostate Magnetic Resonance Imaging to Detect Cancer: Histopathological Correlation Using Prostatectomy Specimens Processed in Customized Magnetic Resonance Imaging Based Molds. J Urology. 2011;186(5):1818–1824. doi: 10.1016/j.juro.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habchi H, Bratan F, Paye A, Pagnoux G, Sanzalone T, Mege-Lechevallier F, Crouzet S, Colombel M, Rabilloud M, Rouviere O. Value of prostate multiparametric magnetic resonance imaging for predicting biopsy results in first or repeat biopsy. Clin Radiol. 2014;69(3):e120–128. doi: 10.1016/j.crad.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 6.Kwak JT, Hewitt SM, Sinha S, Bhargava R. Multimodal microscopy for automated histologic analysis of prostate cancer. BMC Cancer. 2011;11:62. doi: 10.1186/1471-2407-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3(108):108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 8.Doyle S, Feldman MD, Shih N, Tomaszewski J, Madabhushi A. Cascaded discrimination of normal, abnormal, and confounder classes in histopathology: Gleason grading of prostate cancer. BMC Bioinformatics. 2012;13:282. doi: 10.1186/1471-2105-13-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quint L, Van Erp J, Bland P, Del Buono E, Mandell SH, Grossman H, Gikas P. Prostate cancer: correlation of MR images with tissue optical density at pathologic examination. Radiology. 1991;179(3):837–842. doi: 10.1148/radiology.179.3.2028002. [DOI] [PubMed] [Google Scholar]
- 10.Schiebler ML, Tomaszewski JE, Bezzi M, Pollack HM, Kressel HY, Cohen EK, Altman HG, Gefter WB, Wein AJ, Axel L. Prostatic carcinoma and benign prostatic hyperplasia: correlation of high-resolution MR and histopathologic findings. Radiology. 1989;172(1):131–137. doi: 10.1148/radiology.172.1.2472644. [DOI] [PubMed] [Google Scholar]
- 11.Wang XZ, Wang B, Gao ZQ, Liu JG, Liu ZQ, Niu QL, Sun ZK, Yuan YX. Diffusion‐weighted imaging of prostate cancer: Correlation between apparent diffusion coefficient values and tumor proliferation. Journal of Magnetic Resonance Imaging. 2009;29(6):1360–1366. doi: 10.1002/jmri.21797. [DOI] [PubMed] [Google Scholar]
- 12.Zelhof B, Pickles M, Liney G, Gibbs P, Rodrigues G, Kraus S, Turnbull L. Correlation of diffusion-weighted magnetic resonance data with cellularity in prostate cancer. BJU Int. 2009;103(7):883–888. doi: 10.1111/j.1464-410X.2008.08130.x. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs P, Liney GP, Pickles MD, Zelhof B, Rodrigues G, Turnbull LW. Correlation of ADC and T2 measurements with cell density in prostate cancer at 3.0 Tesla. Invest Radiol. 2009;44(9):572–576. doi: 10.1097/RLI.0b013e3181b4c10e. [DOI] [PubMed] [Google Scholar]
- 14.Langer DL, van der Kwast TH, Evans AJ, Plotkin A, Trachtenberg J, Wilson BC, Haider MA. Prostate Tissue Composition and MR Measurements: Investigating the Relationships between ADC, T2, K trans, ve, and Corresponding Histologic Features 1. Radiology. 2010;255(2):485–494. doi: 10.1148/radiol.10091343. [DOI] [PubMed] [Google Scholar]
- 15.Turkbey B, Shah VP, Pang Y, Bernardo M, Xu S, Kruecker J, Locklin J, Baccala AA, Jr, Rastinehad AR, Merino MJ. Is apparent diffusion coefficient associated with clinical risk scores for prostate cancers that are visible on 3-T MR images? Radiology. 2011;258(2):488–495. doi: 10.1148/radiol.10100667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P, Wang SJ, Turkbey B, Grant K, Pinto P, Choyke P, Wood BJ, Summers RM. SPIE Medical Imaging. International Society for Optics and Photonics; 2013. A prostate cancer computer-aided diagnosis system using multimodal magnetic resonance imaging and targeted biopsy labels; pp. 86701G–86701G.pp. 86706 [Google Scholar]
- 17.Shah V, Pohida T, Turkbey B, Mani H, Merino M, Pinto PA, Choyke P, Bernardo M. A method for correlating in vivo prostate magnetic resonance imaging and histopathology using individualized magnetic resonance-based molds. Rev Sci Instrum. 2009;80(10) doi: 10.1063/1.3242697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ojala T, Pietikainen M, Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. Ieee T Pattern Anal. 2002;24(7):971–987. [Google Scholar]
- 19.Guo ZH, Li Q, You J, Zhang D, Liu WH. Local directional derivative pattern for rotation invariant texture classification. Neural Comput Appl. 2012;21(8):1893–1904. [Google Scholar]
- 20.Koço S, Capponi C. Machine Learning and Knowledge Discovery in Databases. Springer; 2011. A boosting approach to multiview classification with cooperation; pp. 209–228. [Google Scholar]
- 21.Vapnik VN. The nature of statistical learning theory. Springer-Verlag New York, Inc; New York: 1995. [Google Scholar]
- 22.Bookstein FL. Principal Warps - Thin-Plate Splines and the Decomposition of Deformations. Ieee T Pattern Anal. 1989;11(6):567–585. doi: 10.1109/34.24792. [DOI] [Google Scholar]
- 23.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Muller M. pROC: an open-source package for R and S plus to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalavagunta C, Zhou X, Schmechel SC, Metzger GJ. Registration of in vivo prostate MRI and pseudo-whole mount histology using Local Affine Transformations guided by Internal Structures (LATIS) J Magn Reson Imaging. 2014 doi: 10.1002/jmri.24629. [DOI] [PMC free article] [PubMed] [Google Scholar]


