Abstract
The population history of Aboriginal Australians remains largely uncharacterised. We generated high-coverage genomes for 83 Aboriginal Australians (speakers of Pama-Nyungan languages) and 25 Papuans from the New Guinea Highlands. We find that Papuan and Aboriginal Australian ancestors diversified 25-40 thousand years ago (kya), suggesting early population structure in the ancient continent of Sahul (Australia, New Guinea and Tasmania). However, all studied Aboriginal Australians descend from a single founding population that differentiated ~10-32 kya. We infer a population expansion in northeast Australia during the Holocene (past c.10 kya) associated with limited gene flow from this region to the rest of Australia consistent with the spread of the Pama-Nyungan languages. We estimate that Aboriginal Australians and Papuans diverged from Eurasians 51-72 kya, following a single out of Africa dispersal, and subsequently admixed with different archaic populations. Finally, we report evidence of selection in Aboriginal Australians potentially associated with living in the desert.
During most of the last 100 ky, Australia, Tasmania and New Guinea formed a single continent, Sahul, which was separated from Sunda (the continental landmass including mainland and western island Southeast Asia) by a series of deep oceanic troughs never exposed by changes in sea level. Colonisation of Sahul is thought to have required at least 8-10 sea crossings between islands, potentially constraining the occupation of Australia and New Guinea by earlier hominins1. Recent assessments suggest that Sahul was settled by 47.5-55 kya2,3 (Figure 1). These dates overlap with those for the earliest evidence for modern humans in Sunda4.
Figure 1. Aboriginal Australian and Papuan samples used in this study, as well as archaeological sites and human remains dated to ~40 kya or older in southern Sunda and Sahul.
The stars indicate the centroid location for each sampling group (sample size in parentheses). Publicly available genetic data (see S04) used as a reference panel in this study are shown as squares. Sites with dated human remains are shown as white circles and the archaeological sites as black circles. The associated dates can be found in S03. Grey boundaries correspond to territories defined by the language groups provided by the Australian Institute of Aboriginal and Torres Strait Islander Studies43. Sampled Aboriginal Australians self-identify primarily as: Yidindji and Gungandji from the Cairns region (CAI, 10, see also S02); Yupangati and Thanakwithi from northwest Cape York (WPA, 6), Wangkangurru and Yarluyandi from the Birdsville region (BDV, 10, 9 sequenced at high depth), Barkindji from southeast (RIV, 8); Pilbara area Yinhawangka and Banjima (PIL, 12), Ngaanyatjarra from western central desert (WCD, 13), Wongatha from WA’s northern Goldfields (WON, 11), Ngadju from WA’s southern Goldfields (NGA, 6); and Nyungar from southwest Australia (ENY, 8). Papuans include samples from the locations Bundi (BUN, 5), Kundiawa (KUN, 5), Mendi (MEN, 5), Marawaka (MAR, 5) and Tari (TAR, 5). We generated SNP array data (black stars) for 45 Papuan samples including 24 Koinambe (KOI) and 15 Kosipe (KOS) - described before44 - and 6 individuals with Highland ancestry sampled in Port Moresby (PMO). Lake Carpentaria (LC), which covered a significant portion of the land bridge between Australia and New Guinea 11.5-40 kya and thus potentially acted as a barrier to gene flow, is also indicated. Map data were sourced from the Australian Government, http://www.naturalearthdata.com/ and our research.
The distinctiveness of the Australian archaeological and fossil record has led to the suggestion that the ancestors of Aboriginal Australians and Papuans (“Australo-Papuans” hereafter) left the African continent earlier than the ancestors of present-day Eurasians5. Although some genetic studies support such multiple dispersals from Africa6, others favour only one out of Africa (OoA) event, with one or two independent founding waves into Asia, of which the earlier contributed to Australo-Papuan ancestry7,8. In addition, recent genomic studies have shown that both Aboriginal Australian8 and Papuan9 ancestors admixed with Neanderthal and Denisovan archaic hominins after leaving Africa.
Increased desertification of Australia10 during the last glacial maximum (LGM) 19-26.5 kya impacted the number and density of human populations11. In this context, unique morphological and physiological adaptations have been identified in Aboriginal Australians living in the desert areas today12. In particular, desert groups were suggested to withstand sub-zero night temperatures without showing the increase in metabolic rates observed in Europeans under the same conditions.
At the time of European contact, Aboriginal Australians spoke over 250 distinct languages, two-thirds of which belong to the Pama-Nyungan family and cover 90% of the Australian mainland13. The place of origin of this language family and the effect of its extensive diffusion on its internal phylogenetic structure, have been debated14, but the pronounced similarity among Pama-Nyungan languages, together with shared socio-cultural patterns, have been interpreted as resulting from a recent, mid-Holocene, expansion15. Other changes in the mid-late Holocene (~4 kya) include the introduction of backed blades and the dingo16. It has been suggested that Pama-Nyungan languages, dingoes and backed blades all reflect the same recent migration into Australia17. Although an external origin for backed blades has been rejected, dingoes were certainly introduced, most likely via island Southeast Asia16. A recent genetic study found evidence of Indian gene flow into Australia at the approximate time of these Holocene changes18, suggesting a possible association, while and substantial admixture with Asians and Europeans is well documented in historical times19.
To date, only three Aboriginal Australian whole genome sequences have been described - one deriving from a historical tuft of hair from Australia’s Western Desert8 and two others from cell lines with limited provenance information20. In this study, we report the first extensive investigation of Aboriginal Australian genomic diversity by analysing the high-coverage genomes of 83 Pama-Nyungan-speaking Aboriginal Australians and 25 Highland Papuans.
Dataset
We collected saliva samples for DNA sequencing in collaboration with Aboriginal Australian communities and individuals in Australia (S01). We sequenced genomes at high-depth (average of 60X, range 20X-100X) from 83 Aboriginal Australian individuals widely distributed geographically and linguistically (Figure 1, Extended Data Table 1, S02-S04). Additionally, we sequenced 25 Highland Papuan genomes (38X-53X; S03, S04) from individuals representative of five linguistic groups, and generated genotype data for 45 additional Papuans living or originating in the highlands (Figure 1). These datasets were combined with previously published genomes and SNP array genotype data, including Aboriginal Australian data from Arnhem Land, and from a human diversity cell line panel from the European Collection of Cell Cultures20 (ECCAC, Figure 1, S04).
We explored the extent of admixture in the Aboriginal Australian autosomal gene pool by estimating ancestry proportions with an approach based on sparse nonnegative matrix factorization (sNMF)21. We found that the genomic diversity of Aboriginal Australian populations is best modelled as a mixture of four main genetic ancestries that can be assigned to four geographic regions based on their relative frequencies: Europe, East Asia, New Guinea and Australia (Figure 2a, Extended Data Figure 1, S05). The degree of admixture varies among groups (S05) with the Ngaanyatjarra speakers from central Australia (WCD) having a significantly higher “Aboriginal Australian component” (median value = 0.95) in their genomes than the other groups sampled (median value = 0.64; Mann-Whitney rank sum test, one tail p-value = 3.55e-07). The “East Asian” and “New Guinean” components are mostly present in northeastern Aboriginal Australian populations, while the “European component” is widely distributed across groups (Figure 2a, Extended Data Figure 1, S05). In most of the subsequent analyses, we either selected specific samples or groups according to their level of Aboriginal Australian ancestry, or masked the data for the non-Aboriginal Australian ancestry genomic components (S06).
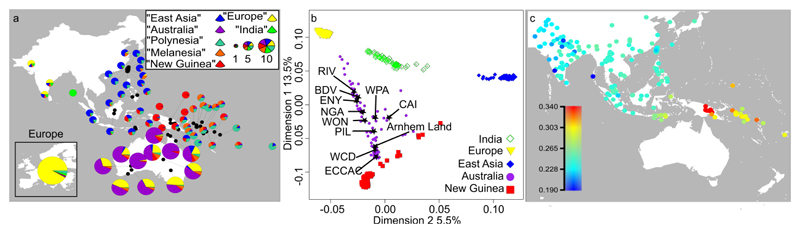
Figure 2. Genetic ancestry of Aboriginal Australians in a worldwide context.
a, Classical Multidimensional scaling (MDS) plot of first two dimensions based on an identity-by-state (IBS) distance matrix (based on 54,971 SNPs) between individuals from this study and worldwide populations, including publicly available data9,18,26,45. The first two dimensions explain 19% of the variance in the IBS distance matrix. Individuals are colour-coded according to sampling location, grouped into Australia (Arnhem Land, ECCAC, BDV, CAI, ENY, NGA, PIL, RIV, WCD, WON, WPA); East Asia (Cambodian, Dai, Han, Japanese, Naxi); Europe (English, French, Sardinian, Scottish, Spanish); India (Vishwabrahmin, Dravidian, Punjabi, Guaharati); and New Guinea (HGDP Papuan, Central Province, Eastern Highlands, Gulf Province, Highlands, PMO, KOI, KOS, BUN, KUN, MEN, TAR, MAR). Stars indicate the centroid for each Aboriginal Australian group. Aboriginal Australians from this study as well as from previous studies are closest to Papuans and also show signals of admixture with Eurasians (see S05 for details). b, Estimation of genomic ancestry proportions for the best number of ancestral components (K=7) based on Aboriginal Australian and Papuan whole genome sequence and SNP array data from this study (see Figure 1), and publicly available SNP array data9,18,26,45 (S05). Each ancestry component has been labelled according to the geographic region showing the corresponding highest frequency. The area of each pie chart is proportional to the sample size (as depicted in the legend). The genomes of Aboriginal Australian populations are mostly a mixture of European and Aboriginal Australian ancestry components. Northern Aboriginal Australian groups (Arnhem Land, CAI, ECCAC, PIL and WPA) are also assigned to components mainly present in East Asian populations, while northeastern Aboriginal Australian groups (CAI and WPA) also show components mainly present in New Guinean populations. A background of 5% “Melanesian” component is observed in all the Aboriginal Australian populations; however, this component is widely spread over the geographic area shown in this figure, being present from Taiwan to India. We detected on average 1.5% “Indian” component and 1.4% “Polynesian” component across the Aboriginal Australian samples, but we attribute these residual ancestry components to statistical noise as they are present in other southeast Asian populations and are not supported by other analyses (S05). c, A heat map displaying outgroup f3 statistics of the form f3(Mbuti; WCD02, X), quantifying genetic drift shared between the putatively unadmixed individual WCD02 chosen to represent the Aboriginal Autralian population, and various populations throughout the broader region for which either array genotypes or whole-genome sequencing data were publicly available or generated in this study. We used 760,116 SNPs for which WCD02 had non-missing array genotypes that overlapped with any other datasets. Standard errors as estimated from block jackknife resampling across the genome were in the range 0.00213-0.00713.
Colonisation of Sahul
The origin of Aboriginal Australians is a source of much debate, as is the nature of the relationships among Aboriginal Australians, and between Aboriginal Australians and Papuans. Using f3 statistics22, estimates of genomic ancestry proportions and classical multidimensional scaling (MDS) analyses, we find that Aboriginal Australians and Papuans are closer to each other than to any other present-day worldwide population considered in our study (Figure 2b, c, S05). This is consistent with Aboriginal Australians and Papuans originating from a common ancestral population which initially colonised Sahul. Moreover, outgroup f3 statistics do not reveal any significant differences between Papuan populations (Highland Papuan groups and HGDP-Papuans) in their genetic affinities to Aboriginal Australians (Extended Data Figure 2a), suggesting that Papuans share a common ancestor after or at the same time as the divergence between Aboriginal Australians and Papuans.
To investigate the number of founding waves into Australia, we contrasted alternative models of settlement history through a composite likelihood method that compares the observed joint Site Frequency Spectrum (SFS) to that predicted under specific demographic models23 (Figure 3, S07). We compared HGDP-Papuans to four Aboriginal Australian population samples with low levels of European admixture (Extended Data Figure 1) from both northeastern (CAI and WPA) and southwestern desert (WON and WCD) Australia. We compared one and two-wave models, where each Australian region was either colonised independently, or by descendants of a single Australian founding population after its divergence from Papuans. The one-wave model is a better fit to the observed SFS, suggesting that the ancestors of the sampled Aboriginal Australians diverged from a single ancestral population. This conclusion is also supported by MDS analyses (Figure 2b), as well as by estimation of ancestry proportion where all Aboriginal Australians form a cluster distinct from the Papuan populations (Extended Data Figure 1, S05). Additionally, it is supported by outgroup f3 analyses, where all Aboriginal Australians are largely equidistant from Papuans when adjusting for recent admixture (Extended Data Figure 2b). Thus, our results, based on 83 Pama-Nyungan speakers, do not support earlier claims of multiple ancestral migrations into Australia giving rise to contemporary Aboriginal Australian diversity24.
Figure 3. Settlement of Australia.
Best supported demographic model of the colonisation of Australia and New Guinea. The demographic history of Aboriginal Australian populations was modelled by considering that sampled individuals are from sub-populations (“islands”) that are part of two larger regions (“continents”), which geographically match the northeast (ne) and the southwestern desert (swd) regions of Australia. Maximum likelihood parameter estimates (MLEs) were obtained from the joint site frequency spectrum (SFS) of Han Chinese, HGDP-Papuans, CAI, WPA, WON and WCD. The 95% CI, obtained by non-parametric block bootstrap, are shown within square brackets. Estimated migration rates scaled by the effective population size (2Nm) are shown above/below the corresponding arrows. Only Aboriginal Australian individuals with low European ancestry were included in this analysis. In this model, we estimated parameters specific to the settlement of Australia and New Guinea (numerical values shown in black); keeping all the other demographic parameters set to the point estimates shown in Figure 4 (numerical value shown in grey here). Only admixture events involving proportions >0.5% are shown. The inferred parameters were scaled using a mutation rate of 1.25x10-8/generation/site40 and a generation time of 29 years corresponding to the average hunter-gatherer generation interval for males and females41. See S07 for further details.
The SFS analysis indicates that there was a bottleneck in the ancestral Australo-Papuan population ~50 kya (95% CI 35-54 kya, S07), which overlaps with archaeological evidence for the earliest occupation of both Sunda and Sahul 47.5-55 kya2,3. We further infer that the ancestors of Pama-Nyungan speakers and Highland Papuans diverged ~37 kya (95% CI 25-40 kya, Figure 3, S07), which is in close agreement with results from MSMC analyses (Extended Data Figure 2c, S08), a method estimating cross coalescence rates between pairs of populations based on individuals’ haplotypes25. It is also in agreement with previous estimates, e.g., based on SNP array data18.
Archaic admixture
We characterised the number, timing and intensity of archaic gene flow events using three complementary approaches: SFS-based (S07), a goodness-of-fit analysis combining D-statistics (S09), and a method that infers putatively derived archaic ‘haplotypes’ (S10). Aboriginal Australians and Papuan genomes show an excess of putative Denisovan introgressed sites (Extended Data Figure 3a, S11), as well as substantially more putative Denisovan-derived haplotypes (PDHs) than other non-Africans (Extended Data Figure 3b, S10). The number and total length of those putative haplotypes vary considerably across samples. However, the estimated number of PDHs correlates almost perfectly (r2 = 0.96) with the estimated proportion of Australo-Papuan ancestry in each individual (Extended Data Figure 3c). We found no significant difference in the distribution of the number of PDHs or the average length of PDHs between putatively unadmixed Aboriginal Australians and Papuans (Mann-Whitney U test, p>0.05). Moreover, the genetic differentiation between WCD and Papuans was also similar for both autosomal SNPs and PDHs with FST values around 0.12. Taken together, these observations provide evidence for Denisovan admixture predating the population split between Aboriginal Australians and Papuans (see also26) and widespread recent Eurasian admixture in Aboriginal Australians (Figure 2a-b, S05). By constraining Denisovan admixture to have occurred before the Aboriginal Australian-Papuan divergence, the SFS-based approach results in an admixture estimate of ~4.0% (95% CI 3.3-5.0%, Figure 4, S07), similar to that obtained by D-statistics (~5%, S09). The SFS analyses further suggest that Denisovan/Australo-Papuan admixture took place ~44 kya (95% CI 31-50 kya, S07).
Figure 4. Out of Africa.
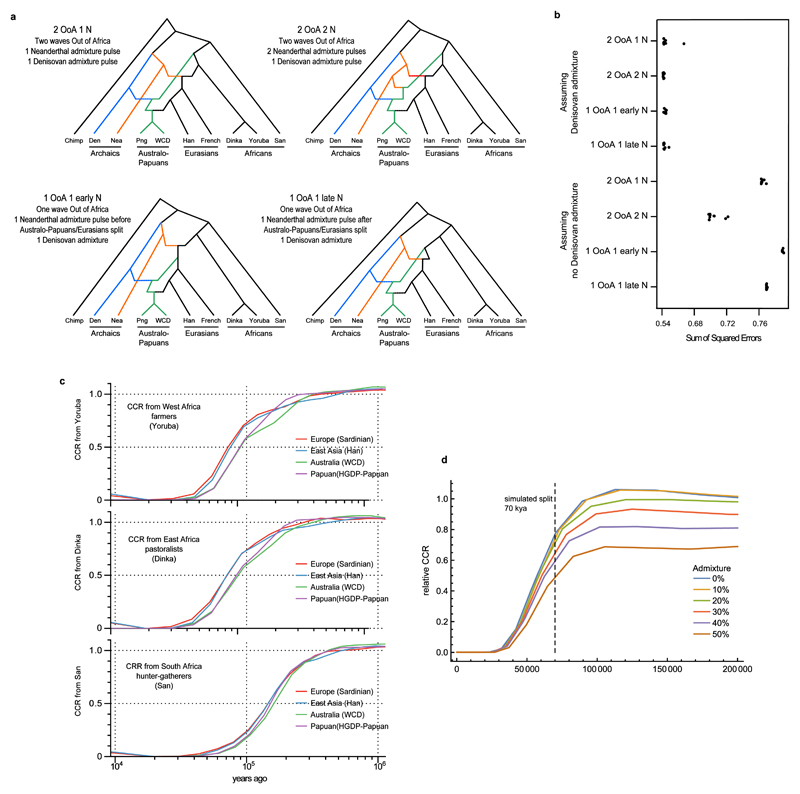
We used a likelihood-based approach to investigate whether the joint SFS supports the one-wave (1 OoA) or two-waves (2 OoA) scenarios. The maximum likelihood estimates (MLEs) are indicative of which scenario is best supported. As shown on the top left inset, under the 1 OoA scenario we expect (i) the presence of an ancestral bottleneck (in black), (ii) a relatively large Neanderthal admixture pulse shared by the ancestors of all non-Africans, and (iii) overlapping divergence times of the ancestors of Aboriginal Australians and Eurasians. In contrast, the top right inset shows parameters expected under a 2 OoA scenario: (i) a limited/absent ancestral bottleneck (in black) in the ancestors of all non-Africans, (ii) no shared Neanderthal admixture in the ancestors of all non-Africans (iii) distinct divergence times for Aboriginal Australians and Eurasians. The main population tree shows the best fitting topology, which supports the 1 OoA scenario, and maximum likelihood estimates (MLEs) for the divergence and admixture times and the admixture proportions (with 95% CI obtained by non-parametric block bootstrap shown within square brackets). We assume that the OoA event is associated with the ancestral bottleneck. The “Ghost” population represents an unsampled population related to Yoruba that is the source of the out of Africa event(s). Our results suggest that these two African populations split significantly earlier (~125 kya) than the estimated time of dispersals into Eurasia. Note that under a 1 OoA scenario, this “Ghost” population becomes, after the ancestral bottleneck, the ancestral population of all non-Africans that admixed with Neanderthals. Arrow thicknesses are proportional to the intensity of gene flow and the admixture proportions, and that only admixture events involving proportions >0.5% are displayed. The inferred parameters were scaled as for Figure 3. See S07 for further details.
The SFS analysis also provides evidence for a primary Neanderthal admixture event (~2.3%, 95% CI 1.1-3.5%, Figure 4, S07) taking place in the ancestral population of all non-Africans ~60 kya (95% CI 55-84 kya, Figure 4, S07). Although we cannot estimate absolute dates of archaic admixture from the lengths of PDHs and putative Neanderthal-derived haplotypes (PNHs) in our samples, we can obtain a relative date. We found that for putatively unadmixed Aboriginal Australians and HGDP-Papuans, the average PNH and PDH lengths are 33.8 kb and 37.4 kb, respectively (Extended Data Figure 3b). These are significantly different from each other (p = 9.65 x10-6 using a conservative sign test), and suggest that the time since Neanderthal admixture was about 11% greater than the time since Denisovan admixture, roughly in line with our SFS based estimates for the Denisovan pulse (31-50 kya) versus the primary pulse of Neanderthal admixture (55-84 kya). The SFS analysis also indicates that the main Neanderthal pulse was followed by a further 1.1% (95% CI 0.2-2.7%, Figure 4, S07) pulse of Neanderthal gene flow into the ancestors of Eurasians. Finally, using our SFS and haplotype based approaches, we explored additional models involving complex structure among the archaic populations. We found suggestive evidence that the archaic contribution could be more complex than a model involving the discrete Denisovan and Neanderthal admixture pulses shown in Figure 48,9 (S07, S10).
Out of Africa
To investigate the relationship of Australo-Papuan ancestors with other world populations, we computed D-statistics22 of the form ((H1=Aboriginal Australian,H2=Eurasian), H3=African) and ((H1=Aboriginal Australian,H2=Eurasian), H3=Ust’-Ishim). Several of these were significantly positive (S09), suggesting that Africans and Ust’-Ishim - a ~45 kya modern human from Asia27 - are both closer to Eurasians than to Aboriginal Australians. These findings are in agreement with a model of Eurasians and Australo-Papuan ancestors dispersing from Africa in two independent waves. However, when correcting for a moderate amount of Denisovan admixture, Aboriginal Australians and Eurasians become equally close to Ust’-Ishim, as expected in a single OoA scenario (S09). Similarly, the D-statistics for ((H1=Aboriginal Australian,H2=Eurasian), H3=African) became much smaller after correcting for Denisovan admixture. Additionally, a goodness-of-fit approach combining D-statistics across worldwide populations indicates stronger support for two waves OoA, but when taking Denisovan admixture into account, a one-wave scenario fits the observed D-statistics equally well (Extended Data Figure 4a-b, S09).
To further investigate the timing and number of OoA events giving rise to present-day Australo-Papuans and Eurasians we used the observed SFS in a model-based composite likelihood framework. When considering only modern human genomes, we find evidence for two waves OoA, with a dispersal of Australo-Papuans ~14 ky before Eurasians (S07). However, when explicitly taking into account archaic Neanderthal and Denisovan introgression into modern humans9,20, the SFS analysis supports a single origin for the OoA populations marked by a bottleneck ~72 kya (95% CI 60-104 kya, Figure 4, S07). This scenario is reinforced by the observation that the ancestors of Australo-Papuans and Eurasians share a 2.3% (95% CI 1.1-3.5%) Neanderthal admixture pulse. Furthermore, modern humans have both an LD decay rate and a number of predicted deleterious homozygous mutations (recessive genetic load) that correlates with distance from Africa (S05, S11, Extended Data Figure 5), again consistent with a single African origin. The model estimated from the SFS analysis also suggests an early divergence of Australo-Papuans from the ancestors of all non-Africans, in agreement with two colonisation waves across Asia8,9,18. Under our best model, Australo-Papuans began to diverge from Eurasians ~58 kya (95% CI 51-72 kya, Figure 4, S07), whereas Europeans and East Asians diverged from each other ~42 kya (95% CI 29-55 kya, Figure 4, S07), in agreement with previous estimates7,18,28. We find evidence for high levels of gene flow between the ancestors of Eurasians and Australo-Papuans, suggesting that, after the fragmentation of the OoA population (“Ghost” in Figure 4) 57-58 kya, the groups remained in close geographical proximity for some time before Australo-Papuan ancestors dispersed eastwards. Furthermore, we infer multiple gene flow events between sub-Saharan Africans and Western Eurasians after ~42 kya, in agreement with previous findings of gene flow between African and non-African populations28.
MSMC analyses suggest that the Yoruba/Australo-Papuans and the Yoruba/Eurasians cross-coalescence rates are distinct, implying that the Yoruba and Eurasian gene trees across the genome have, on average, more recent common ancestors (Extended Data Figure 4c, S08). We show through simulations that these differences cannot be explained by typical amounts of archaic admixture (<20%, Extended Data Figure 4d). Moreover, the expected difference in phasing quality among genomes is not sufficient to fully explain this pattern (S08). While a similar separation in cross coalescence rate curves is obtained when comparing Eurasians and Australo-Papuans with Dinka, we find that, when comparing Australo-Papuans and Eurasians with San, the cross coalescence curves overlap (Extended Data Figure 4c). We also find that the inferred changes in effective population size through time of Aboriginal Australians, Papuans, and East Asians are very similar until around 50 kya, including a deep bottleneck around 60 kya (Extended Data Figure 6). Taken together, these MSMC results are consistent with a split of both Australo-Papuans and Eurasians from a single African ancestral population, combined with gene flow between the ancestors of Yoruba or Dinka (but not San) and the ancestors of Eurasians that is not shared with Australo-Papuans. These results are qualitatively in line with the SFS-based analyses (see e.g., Figure 4). While our results do not exclude the possibility of an earlier OoA expansion, they do indicate that any such event left little or no trace in the genomes of modern Australo-Papuans.
Genetic structure of Aboriginal Australians
Uniparental haplogroup diversity in this dataset (Extended Data Table 1, S12) is consistent with previous studies of mitochondrial DNA (mtDNA) and Y chromosome variation in Australia and Oceania29, including the presence of typically European, Southeast and East Asian lineages30. The combined results provide important insights into the social structure of Aboriginal Australian societies. Aboriginal Australians exhibit greater between-group variation for mtDNA (16.8%) than for the Y chromosome (11.3%), in contrast to the pattern for most human populations31. This result suggests higher levels of male- than female-mediated migration, and may reflect the complex marriage and post-marital residence patterns among Pama-Nyungan Australian groups32. As expected (S02), the inferred European ancestry for the Y chromosome is much greater than that for mtDNA (31.8% vs. 2.4%), reflecting male-biased European gene flow into Aboriginal Australian groups during the colonial era.
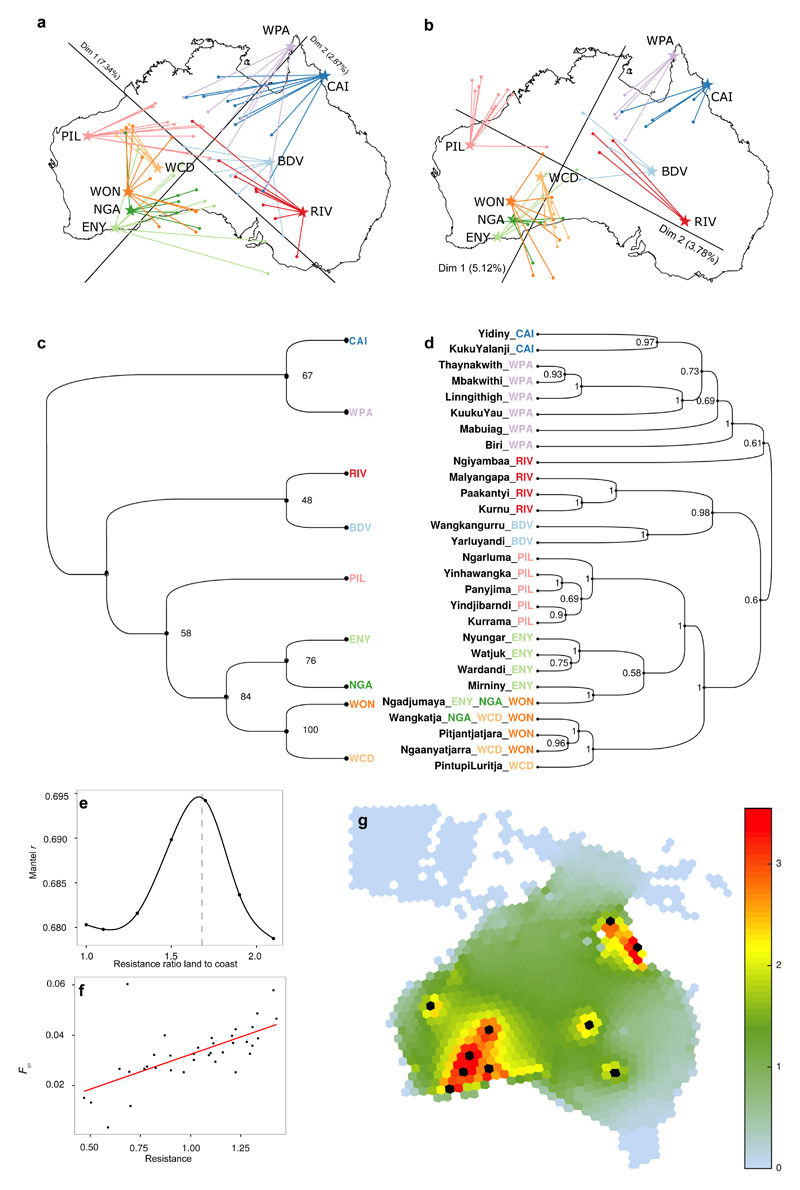
On an autosomal level, we find that genetic relationships within Australia reflect geography, with a significant correlation (rGEN,GEO = 0.77, p-value < 0.0005, Extended Data Figure 7b) between the first two dimensions of an MDS analysis on masked genomes with geographical location (S13). Populations from the centre of the continent occupy genetically intermediate positions (Extended Data Figure 7a-b). A similar result is observed with an FST-based tree for the masked data (Extended Data Figure 7c, S05) as well as in analyses of genetic affinity based on f3 statistics (Extended Data Figure 2a), suggesting a population division between northeastern and southwestern groups. This structure is further supported by SFS analyses showing that populations from southwestern desert and northeastern regions diverged as early as ~31 kya (95% CI 10-32 kya), followed by limited gene flow (estimated 2Nm<0.01, 95% CI 0.00<Nm< 11.25). The analysis of the major routes of gene flow within the continent supports a model in which the Australian interior acted as a barrier to migration. Using a model inspired by principles of electrical engineering where gene flow is represented as a current flowing through the Australian continent and using observed FST values as a proxy for resistance, we infer that gene flow occurred preferentially along the coasts of Australia (Extended Data Figure 7e-g, S13). These findings are consistent with a model of expansion followed by population fragmentation when the extreme aridity in the interior of Australia formed barriers to population movements during the LGM33.
We used MSMC on autosomal data and mtDNA Bayesian Skyline Plots34 (BSP) to estimate changes in effective population size within Australia. The MSMC analyses provide evidence of a population expansion starting ~10 kya in the northeast, while both MSMC and BSP indicate a bottleneck in the southwestern desert populations taking place during the past ~10 kya (Extended Data Figure 6, S08, S12). This is consistent with archaeological evidence for a population expansion associated with significant changes in socio-economic and subsistence strategies in the Holocene35 that impacted Australia.
European admixture almost certainly had not occurred before the late 18th century, but earlier East Asian and/or New Guinean gene flow into Australia could have taken place. We characterised the mode and tempo of gene flow into Aboriginal Australians using three different approaches (S06, S07, S14). We used approximate Bayesian computation (ABC) to compare the observed mean and variance in the proportion of European, East Asian and Papuan admixture among Aboriginal Australian individuals, to that computed from simulated datasets under various models of gene flow. We estimated European and East Asian admixture to have occurred on the order of ten generations ago (S14), consistent with historical and ethnographic records. Consistent with this, the local ancestry approach suggests that European and East Asian admixture is more recent than Papuan admixture (Extended Data Figure 4a, S06). In addition, both ABC and SFS analyses indicate that the best-fitting model for the Aboriginal Australian-Papuan data is one of continuous but modest gene flow, mostly unidirectional from Papuans to Aboriginal Australians, and geographically restricted to northeast Aboriginal Australians (2Nm=0.41, 95% CI 0.00-20.35, Figure 3, S07).
To further investigate gene flow from New Guinea, we conducted analyses on the Papuan ancestry tracts obtained from the local ancestry analysis. We inferred local ancestry as the result of admixture between four components: European, East Asian, Papuan and Aboriginal Australian (S06). Papuan tract length distribution shows a clear geographic pattern (Extended Data Figure 8); we find a strong correlation of Papuan tract length variance with distance from WCD to other Aboriginal Australian groups (r=0.64, p-value<0.0001). The prevalence of short ancestry tracts of Papuan origin, compared to longer tracts of East Asian and European origin, suggests that a large fraction of the Papuan gene flow is much older than that from Europe and Asia, which is consistent with the ABC analysis (S14). We also investigated possible South Asian (Indian related) gene flow into Aboriginal Australians, as reported recently18. However, we found no evidence of a component that can be uniquely assigned to Indian populations in the Aboriginal Australian gene pool using either admixture analyses or f3 and D-statistics (S05), even when including the original Aboriginal Australian genotype data from Arnhem Land. The different size and nature of the comparative datasets may account for the discrepancy in the results.
Pama-Nyungan languages and genetic structure
To investigate whether or not linguistic relationships reflect genetic relationships among Aboriginal Australian populations, we built a Bayesian phylogenetic tree for the 28 different Pama-Nyungan languages represented in this sample13 (Extended Data Table 1, S15). The resulting linguistic and FST-based genetic trees (Extended Data Figure 7c,d) share several well-supported partitions. For example, both trees indicate that the northeastern (CAI and WPA) and southwestern groups (ENY, NGA, WCD and WON) form two distinct clusters, while PIL, BDV and RIV are intermediate. A distance matrix between pairs of languages, computed from the language-based tree, is significantly correlated with geographic distances (rGEO,LAN = 0.83, Mantel test two-tail p-value on 9,999 permutations = 0.0001, S13). This suggests that differentiation among Pama-Nyungan languages in Australia follows geographic patterns, as observed in other language families elsewhere in the world6. Furthermore, we find a correlation between linguistics and genetics (rGEN,LAN= 0.43, Mantel test p-value < 0.0005, S13) that remains significant when controlling for geography (rGEN,LAN.GEO= 0.26, partial Mantel test p-value < 0.0005, S13). This is consistent with language differentiation after populations loose (genetic) contact with one another. The correlation between the linguistic and genetic trees is all the more striking given the difference in time scales: the Pama-Nyungan family is generally accepted to have diversified within the last 6 ky36, while the genetic estimates are two to five times that age. The linguistic tree thus cannot simply reflect initial population dispersals, but rather reflects a genetic structure that has a complex history, with initial differentiation 10-32 kya, localised population expansions (northeast) and bottlenecks (southwest) ~10 kya, and subsequent limited gene flow from the northeast to the southwest. The latter may be the genetic signature that tracks the divergence of the Pama-Nyungan language family.
Selection in Aboriginal Australians
To identify selection signatures specific to Aboriginal Australians, we used two different methods based on the identification of SNPs with high allele frequency differences between Aboriginal Australians and other groups, similar to the Population-Branch Statistics37 (PBS, S16). First, we scanned the Aboriginal Australian genomes for loci with unusually large changes in allele frequency since divergence from Papuans, taking recent admixture with Europeans and Asians into account (“global scan”). Second we identified genomic regions showing high differentiation associated with different ecological regions within Australia (“local scan”, S16). Among the top ranked peaks (Extended Data Table 2) we found genes associated with the thyroid system (NETO1, 7th peak in the global scan, and KCNJ2, 1st peak in the local scan) and serum urate levels (8th peak in the global scan). Thyroid hormone levels are associated with Aboriginal-Australian-specific adaptations to desert cold38 and elevated serum urate levels with dehydration39. These genes are therefore candidates for potential adaptation to life in the desert. However, further studies are needed to associate putative selected genetic variants with specific phenotypic adaptations in Aboriginal Australians.
Discussion
Australia has one of the longest histories of continuous human occupation outside Africa, raising questions of origins, relatedness to other populations, differentiation and adaptation. Our large scale genomic data and analyses provide some answers but also raise new questions. We find that Aboriginal Australians and Eurasians share genomic signatures of an OoA dispersal - a common African ancestor, a bottleneck and a primary pulse of Neanderthal admixture. However, Aboriginal Australian population history diverged from that of other Eurasians shortly after the OoA event, and included private admixture with another archaic hominin.
Our genetic-based time estimates are relative, and to obtain absolute dates we relied on two rescaling-parameters: the human mutation rate and generation time (assumed to be 1.25×10-8/generation/site and 29 years, respectively, based on recent estimates40,41). Although the absolute estimates we report would need to be revised if these parameters were to change, the current values can be the starting point of future research and should be contextualized.
We find a relatively old divergence between the ancestors of Pama-Nyungan speakers and Highland Papuans, only ~10% younger than the European-East Asian split time. With the assumed rescaling-parameters this corresponds to ~37 kya (95% CI 25-40 kya) implying that the divergence between sampled Papuans and Aboriginal Australians is older than the disappearance of the land bridge between New Guinea and Australia about 7-14.5 kya, and thus suggests ancient genetic structure in Sahul. Such structure may be related to palaeo-environmental changes leading up to the LGM. Sedimentary studies show that the large Lake Carpentaria (500 x 250 km, Figure 1) formed ~40 kya, when sea-levels fell below the 53 m-deep Arafura Sill42. Although Australia and New Guinea remained connected until the early Holocene, the flooding of the Carpentaria basin and its increasing salinity46 24 45may have thus promoted population isolation.
Our results also indicate that the population that diverged from Papuans ~37 kya was ancestral to all Aboriginal Australian groups sampled in this study; yet, archaeological evidence shows that by 40-45 kya, humans were widespread within Australia (Figure 1). Three non-exclusive scenarios could account for this observation: 1) the Aboriginal Australian ancestral population was widespread prior to the divergence from Papuans, maintaining gene flow across the continent; 2) it was deeply structured, and only one group survived to give rise to modern Aboriginal Australians; and 3) other groups survived, but the descendants are not represented in our sample. Additional modern genomes, especially from Tasmania and the non-Pama-Nyungan regions of the Northern Territory and Kimberley, as well as ancient genomes pre-dating European contact in Australia and other expansions across Southeast Asia17, may help resolve these questions in the future.
Data access
The Aboriginal Australian whole genome sequence data and the SNP array data generated in this study are available upon request by contacting the research ethics and integrity service at Griffith University (rick.williams@griffith.edu.au). The Papuan whole genome sequence data generated in this study are available under managed access through the EGA database (https://www.ebi.ac.uk/ega) under study accession number EGAS00001001247.
Extended Data
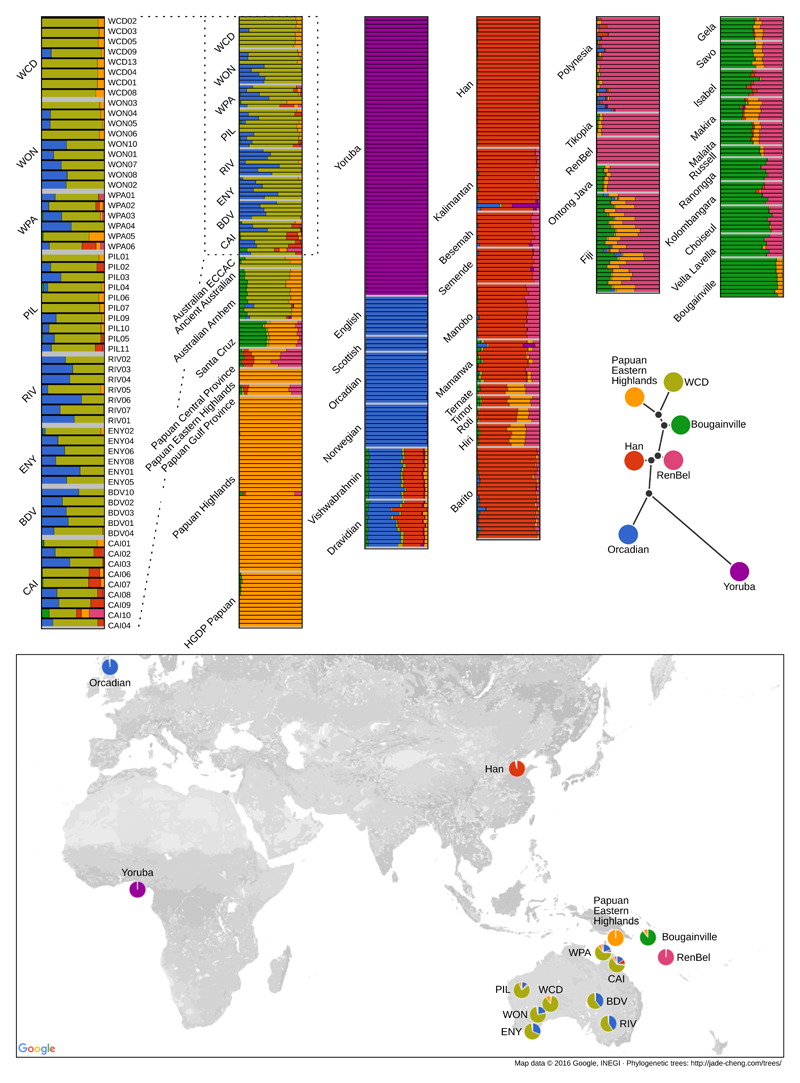
Extended Data Figure 1. Per individual admixture proportions of K=7 ancestral components including Aboriginal Australians, New Guineans, Europeans, Africans, Melanesians and Polynesians.
The genome of each individual is depicted as a bar and is coloured according to the estimated genome-wide proportions of ancestry components. An unrooted tree showing the relationships between the identified ancestral components is also estimated by our method. Each ancestry has been labelled with the name of the population (see also map) showing the highest fraction of that ancestral component. The cross validation (CV) error is minimised for this value of K for 5-fold CV (S05). The rooted tree supports the shared genetic origin of Aboriginal Australians, Papuans and Bougainvilleans.
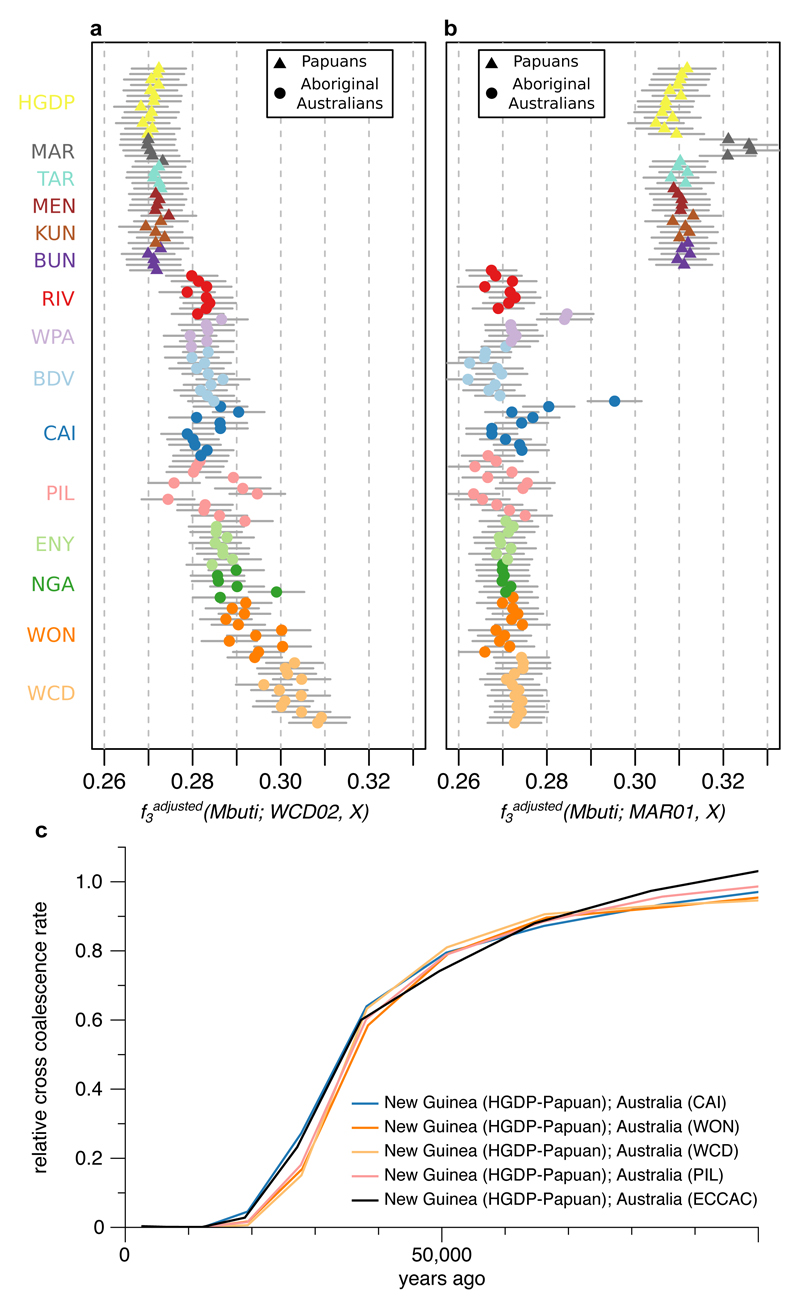
Extended Data Figure 2. Genetic relationships of Aboriginal Australians and Papuans.
a, Genetic affinities between a Western Central Desert (WCD02) genome and Aboriginal Australians and Papuans. Outgroup f3 statistics between WCD02 and all other Aboriginal Australians and Highland Papuan individuals that were whole-genome sequenced for this study, using all genotypes called from the sequencing data. Because the widespread recent admixture in Aboriginal Australians has large confounding effects on the f3 statistics, the values were adjusted using the slope coefficient from a simple linear regression model fitted to the relationship between f3 and the fraction of non-indigenous (i.e., not Aboriginal Australian nor Papuan) ancestry in each individual genome. The adjusted f3 statistics display a genetic gradient that separates western and eastern Aboriginal Australian populations. However, we find no differences between Papuan population samples in their level of Aboriginal Australian affinity (Kruskal-Wallis test, p-value = 0.083). Horizontal lines correspond to ±1 standard error. b, Genetic affinities between a Papuan highlander genome and Aboriginal Australians and Papuans. The Papuan highlander sample MAR01 from the Marawaka area was arbitrarily chosen as a reference point for this analysis. f3 values were adjusted for recent admixture as in (a). All Aboriginal Australian groups display a similar level of Highland Papuan affinity (with the exception of three outlier individuals from the north-eastern WPA and CAI populations: WPA06, WPA05 and CAI10, the latter two of which are known to have at least one parent with origins in Papua New Guinea or the Torres Strait Islands). While some differences between groups are actually statistically significant (Kruskal-Wallis test, p-value = 0.0002, after removing the three outliers), which could be consistent with e.g. low levels of Papuan gene flow into some Aboriginal Australian groups (see S06 and S07), we caution that some of these differences are likely due to imperfect adjustment for Eurasian admixture (the adjusted f3 is highest in the WCD population, which has the least Eurasian admixture). Horizontal lines correspond to ±1 standard error. c, MSMC analyses. Linear interpolation through the midpoints of the time intervals of the relative cross coalescence rate estimates from MSMC25 using pairs of individuals including one HGDP-Papuan and one other individual as indicated. We used CAI01, PIL06, WCD01, WON03 and an ECCAC sample for this analysis (see S08 for details). The MSMC results were scaled using a mutation rate of 1.25x10-8 /generation/site as suggested in40 and a generation time of 29 years corresponding to the average hunter-gatherer generation interval for males and females41.
Extended Data Figure 3. Introgressed archaic sites and putative Denisovan and Neanderthal haplotypes.
a, Distribution of per individual number of putative introgressed sites from archaic humans. The number of Neanderthal-specific introgressed sites increases from Europe to Australia, and then decreases in Amerindians, which is consistent with recurrent Neanderthal (or Neanderthal-related archaic) gene flow during the expansion into Eurasia. Our results are thus indicative of several pulses of Neanderthal gene flow into modern humans, as inferred previously46–48. We note, however, that the apparent high levels in Neanderthal-specific introgressed sites in Australo-Papuans can be explained by the expected number of misclassified Neanderthal introgressed sites resulting from the shared ancestry with Denisovans (see S10 for details). b, c, d, e, Putative Denisovan (PDH) and Neanderthal haplotypes (PNH). The putative haplotypes correspond to clusters (four or more SNPs spanning at least 4kb) of heterozygous or homozygous genotypes in complete linkage disequilibrium (“diplotypes”) that are potentially the result of Neanderthal or Denisovan admixture. Those diplotypes are homozygous ancestral in 10 Africans, homozygous derived in the Denisovan for the PDH (respectively Neanderthal for the PNH), homozygous ancestral in the Neanderthal for the PDH (respectively Denisovan for the PNH), and with the derived allele segregating in all other contemporary non-African humans (see S11 for details). We report the average number of the PDHs and PNHs (b), the correlation between the estimated amount of Australo-Papuan ancestry (see Figure 2b, Extended Data Figure 1, S05) and the number of identified PDHs for each Australian sample, the sum of the lengths (d) and the average length (d) of the PDHs and PNHs per individual for worldwide populations included in our reference panel (see S03).
Extended Data Figure 4. Out of Africa: admixture graphs based on D-statistic and MSMC analyses.
a, Admixture graphs representing some of the topologies considered for the two waves and one wave Out of Africa models assuming Denisovan admixture. All topologies are identical except for the coloured lineages representing Australo-Papuans (green), Neanderthal (Nea, orange) and Denisovan (Den, blue). The graphs differ in (i) the number of OoA events, and (ii) the number of Neanderthal admixture pulses. Png stands for HGDP-Papuan. b, Sum of Squared Errors between the observed D-statistics and the expectations for each quartet in the graph involving the chimpanzee as an outgroup for each of the admixture graphs shown in a and the corresponding four without Denisovan admixture. Each point is the result of the optimization procedure with different starting points. See S09 for details. c, MSMC analyses. Relative cross coalescence rate (CCR) estimates from MSMC25 for pairs of individuals including one African sample (Yoruba, Dinka and San) and one other sample from Eurasia, as indicated in the legend. d, Simulation study to assess the effect of archaic admixture on the CCR rates. Relative CCR estimated for data simulated under a simple two population divergence model where one of the populations admixed at different rates with an archaic population. See S08 for details.
Extended Data Figure 5. Inferred deleterious mutations.
a, Boxplot of the number of derived homozygous sites per individual for worldwide populations that are predicted to be deleterious. Deleteriousness of SNPs was inferred using GERP Rejected Substitution (RS) scores. Derived alleles with a RS score larger than 2 were considered to be deleterious, see S11. Average RS score per individual calculated across heterozygous sites (b), and derived homozygous sites (c). Each coloured symbol corresponds to estimates from a single individual. Homozygosity is calculated as the number of derived homozygous sites divided by the number of sites at which an individual carries at least one copy of the derived allele. Solid lines show the linear regression of homozygosity against average RS score per individual for non-African modern humans. Dashed lines indicate the 95% confidence interval for the linear regression.
Extended Data Figure 6. Effective population size changes over time.
a, MSMC analyses. Population size estimates from MSMC for pairs of individuals from several populations within and outside of Australia. For each run, we used two individuals from each population, i.e., four haplotypes in each run. MSMC results were scaled as in Figure 3. b, Bayesian Skyline Plots. Bayesian Skyline Plots (BSP) calculated from the mtDNA genome sequences, showing the effective population size estimates over time when considering either groups from northeastern Australia (CAI, WPA) or groups from southwestern Australia (ENY, NGA, WCD, WON). Solid lines are the estimates, dashed lines are the corresponding 95% credible intervals (see S12).
Extended Data Figure 7. Genetics mirrors geography and languages.
a, b. Procrustes analyses of the first two dimensions of a classical multidimensional scaling (MDS) analysis of the Aboriginal Australian genome sequences (autosomes). We considered two cases: an analysis including all variants (a), or only the variants remaining after genomic regions of putative recent European and East Asian (i.e., Han Chinese) origin are “masked” (b, S06). Both MDS plots have been rotated towards the best overlap with geographic sampling locations as defined by Procrustes analysis49. In each plot, the arrows indicate the error of the MDS coordinates towards the assigned population sampling geographic coordinates. We find that the genetic relationships within Australia mirrors geography, with a significant correlation for both cases, i.e. rGEN,GEO = 0.59, p-value < 0.0005 for all variants and even higher (rGEN,GEO = 0.77, p-value < 0.0005) for the masked data. We find using the Bearing correlogram approach that the main axis of genetic differentiation in the masked Aboriginal Australian genomes is at angle = 65° compared to the equator, i.e., in the southwest to northeast direction (S13). c,d. Correspondence between genetics and linguistics. Unrooted Neighbor-Joining FST-based genetic tree (cladogram). Weir and Cockerham FST distance was computed between the Aboriginal Australian populations after masking the Eurasian tracts. Statistical robustness of each branch was estimated by means of a bootstrap analysis (1000 replicates, S05). d, Bayesian phylogenetic tree for the 28 different Pama-Nyungan languages represented in this sample (from13, see S15). Posterior probabilities are also indicated. Note that one language group can be shared by different Aboriginal Australian groups. The linguistic tree was built with BEAST (50). e,f,g. Gene flow across the continent. e, Mantel non-parametric r (estimating the goodness of fit between genetic differentiation and connectivity) versus ratios of resistance of inland to coastal nodes, showing a peak at 1.7. f, Best fit of pairwise population genetic differentiation, FST (computed between the nine Aboriginal Australian groups after masking Eurasian tracts (S06)), versus pairwise connectivity based on the environment (estimated as resistance) when moving inland is 1.7 times harder than moving along coastal nodes. g, Gene flow across the Australian landscape, quantified as the cumulative current for pairwise connections among Aboriginal Australian groups (black circles), with larger current (warmer colours) representing greater gene flow.
Extended Data Figure 8. European, East Asian and Papuan genomic tracts in Aboriginal Australians.
a, Distribution of the tracts assigned to Aboriginal Australian (WCD), Papuan, East Asian or European ancestry for 58 unrelated non-WCD Australian samples. Most of the shorter tracts were of Papuan origin, suggesting that a large fraction of the Papuan gene flow is much older than that from Europe and East Asia, consistent with a Papuan influence spreading slowly from northeastern to southwestern Australia by ancient migration. b, Corresponding scatter plot with fitted line of per-individual variance in Papuan tract length vs. geographic distance from WCD, the latter calculated using the Great Circle Distance formula for pairs of individual GPS coordinates. Papuan tract distribution showed a strong and significant correlation with distance from WCD (r = 0.64; p-value < 10-5), with “younger tracts” closer to New Guinea (i.e., with a larger variance) and “older tracts” closer to WCD” (i.e., with a smaller variance). This is also consistent with continuous Papuan gene flow spreading from the northeast.
Extended Data Table 1. Whole genome sequence depth of coverage, haplogroup and language assignments for the Aboriginal Australian samples.
| indiv. | DoC* | mtDNA haplotype† | Ychr haplotype‡ | Pama-Nyungan language§ |
|---|---|---|---|---|
| BDV01 | 78 | S2 | - | Yarluyandi Wangkangurru |
| BDV02 | 75 | S1a | R1b1a2a1a2c1g2a1a2 | Yarluyandi Wangkangurru |
| BDV03 | - | - | - | Yarluyandi Wangkangurru |
| BDV04 | 70 | O1a | - | Yarluyandi Wangkangurru |
| BDV05 | 72 | S1a | O1a | Yarluyandi Wangkangurru |
| BDV06 | 70 | S1a | - | Yarluyandi Wangkangurru |
| BDV07 | 70 | O1a | - | Yarluyandi Wangkangurru |
| BDV08 | 70 | S1a | R1b1a2a1a2c1g2a1 | Yarluyandi Wangkangurru |
| BDV09 | 74 | S1a | - | Yarluyandi Wangkangurru |
| BDV10 | 72 | S1a | I1a2a1d | Yarluyandi Wangkangurru |
| CAI01 | 84 | P | K2b | Yidiny |
| CAI02 | 74 | M42 | K2b | Yidiny |
| CAI03 | 77 | M42a | - | Yidiny |
| CAI04 | 71 | P | - | Yidiny KukuYalanji |
| CAI05 | 80 | P | O2a1a | Yidiny |
| CAI06 | 78 | P | C1b | Yidiny |
| CAI07 | 71 | N13 | K2b | KukuYalanji |
| CAI08 | 70 | P | K2b | Yidiny |
| CAI09 | 79 | P | R1b1a2a1a2b1 | Yidiny |
| CAI10 | 73 | E1a2 | K2b | - |
| ENY01 | 69 | H1e1a3 | R1b1a2a1a2b1c1 | Nyungar |
| ENY02 | 79 | R12 | - | Ngadjumaya |
| ENY03 | 83 | O | - | Mirniny |
| ENY04 | 83 | M42 | - | Nyungar |
| ENY05 | 78 | S2 | - | Ngadjumaya |
| ENY06 | 70 | M42 | - | Wardandi |
| ENY07 | 73 | S2 | E1b1b1b2a | Watjuk |
| ENY08 | 71 | P4b1 | C1b | Nyungar Ngadjumaya |
| NGA01 | 74 | O1 | - | Ngadjumaya |
| NGA02 | 52 | O1a | - | Ngadjumaya |
| NGA03 | 73 | O | - | Ngadjumaya |
| NGA04 | 75 | O | R1b1a2a1a1b1a1a | Wangkatja |
| NGA05 | 56 | R12 | - | Ngadjumaya |
| NGA06 | 63 | S1a | - | Wangkatja |
| PIL01 | 58 | R | C1b | Yinhawangka |
| PIL02 | 61 | M42 | C1b | Yinhawangka |
| PIL03 | 56 | M42 | - | Yinhawangka |
| PIL04 | 64 | M42 | - | Yinhawangka |
| PIL05 | 68 | M42 | C1b | Yinhawangka |
| PIL06 | 59 | O1 | K2b | Panyjima |
| PIL07 | 63 | O | - | Panyjima |
| PIL08 | 72 | M42 | C1b | Yindjibarndi Kurrama |
| PIL09 | 58 | S5 | R1b1a2a1a2e1 | Kurrama |
| PIL10 | 61 | R | - | Yinhawangka |
| PIL11 | 57 | P3b | C1b | Kurrama |
| PIL12 | 63 | P3b | C1b | Yindjibarndi |
| RIV01 | 73 | M42a | - | Ngiyambaa |
| RIV02 | 62 | P4b1 | - | Paakantyi |
| RIV03 | 69 | M42a | - | Paakantyi |
| RIV04 | 62 | P4b1 | I2a1a2a1a | Kurnu |
| RIV05 | 72 | P4b1 | - | Paakantyi |
| RIV06 | 66 | H1bs | J2a1b | Ngiyambaa |
| RIV07 | 70 | P4b1 | R1b1a2a1a2c1c | Paakantyi |
| RIV08 | 66 | P4b1 | - | Paakantyi Malyangapa |
| WCD01 | 62 | R12 | K2b | Ngaanyatjarra |
| WCD02 | 59 | S1a | C1b | Ngaanyatjarra |
| WCD03 | 61 | R12 | K2b | Wangkatja |
| WCD04 | 52 | P3b | K2b | Ngaanyatjarra |
| WCD05 | 60 | O1 | C1b | Ngaanyatjarra |
| WCD06 | 58 | O1a | C1b | Ngaanyatjarra |
| WCD07 | 61 | M42 | - | Ngaanyatjarra |
| WCD08 | 64 | M42 | - | Ngaanyatjarra |
| WCD09 | 59 | R | J2a1b | Ngaanyatjarra |
| WCD10 | 63 | M42 | - | Ngaanyatjarra |
| WCD11 | 57 | M42 | K2b | Ngaanyatjarra |
| WCD12 | 59 | M42 | C1b | Ngaanyatjarra PintupiLuritja |
| WCD13 | 67 | M14 | C1b | Ngaanyatjarra |
| WON01 | 71 | O | I1a2a1a3a | Wangkatja |
| WON02 | 101 | O1a | - | Wangkatja |
| WON03 | 65 | O1a | - | Wangkatja |
| WON04 | 58 | R | - | Ngaanyatjarra |
| WON05 | 56 | O1a | I2a2a1a2a2 | Wangkatja |
| WON06 | 60 | R12 | - | Wangkatja |
| WON07 | 57 | O | - | Ngadjumaya |
| WON08 | 52 | O | - | Wangkatja |
| WON09 | 20 | O | E1b1b1a1b1a4 | Wangkatja |
| WON10 | 50 | O1 | R1b1a2a1a2a | Wangkatja |
| WON11 | 58 | R12 | - | Pitjantjatjara |
| WPA01 | 51 | P5 | - | Thaynakwith Linngithigh |
| WPA02 | 50 | P | C1b | Mpakwithi Kaanju |
| WPA03 | 51 | M42a | K2b | Thaynakwith Biri |
| WPA04 | 52 | P5 | - | Thaynakwith KukuYau |
| WPA05 | 56 | M42 | NA | Mabuiag Thaynakwith |
| WPA06 | 53 | P5 | O1a | Mpakwithi |
The depth of coverage (DoC) is the average number of reads covering every position in the genome (hg19) after duplicate removal (see S03).
The average depth of coverage on the mitochondrial genome (mtDNA) is 3,484 ± 1,515 (mean ± SD) and haplogroups were called with haplogrep (http://haplogrep.uibk.ac.at/) and haplofind (https://haplofind.unibo.it/), see S12 for details and references.
The average depth of coverage on the Y chromosome (Ychr) is 28.88 ± 4.5 (mean ± SD). Haplogroup assignment was performed with an in-house script that matched our SNPs with the classification provided in ISOGG version 10.08, see S12 for details and references.
Language group with which the speaker self-identifies, or to which they were assigned. Where more than one language is given, speakers either identified with more than one group, or they could not be assigned to a single group with certainty.
Extended Data Table 2. Selection scan in Aboriginal Australians.
| Focal Pop | Nearby Gene* | Position† | rsID | Dist ‡ | PBSn§ | F12 ¶ | F13 | F23 | Function of gene product# |
|---|---|---|---|---|---|---|---|---|---|
| All | TMEM86B | 55,833,076 | rs734517 | 92,444 | 0.78 | 0.93 | 0.99 | 0.06 | Catalyzes the degradation of lysoplasmalogen. Modulates cell membrane proteins. |
| All | LRRC52 | 165,621,695 | rs4147601 | 88,510 | 0.74 | 0.96 | 0.91 | 0.01 | Modulates voltage of potassium ion channels. Expressed in testis. |
| All | MACROD2 | 15,209,684 | rs175279 | 901 | 0.70 | 0.92 | 0.89 | -0.01 | Involved in deacetylase activity. Possibly (but not conclusively) causative of Kabuki syndrome. |
| All | JRKL | 96,747,146 | rs72959058 | 507,105 | 0.74 | 0.99 | 0.87 | 0.15 | Homologue to “jerky” gene in mouse. |
| All | SPATA20 | 48,631,324 | rs73338243 | 287 | 0.70 | 0.96 | 0.85 | 0.09 | Spermatid protein. |
| All | NAA60 | 3,537,933 | rs73503305 | 970 | 0.71 | 0.91 | 0.91 | -0.02 | Histone acetyltransferase required for nucleosome assembly and chromosome segregation during anaphase. Human-specific imprinted gene. |
| All | CBLN2 | 70,019,066 | rs12455116 | 184,848 | 0.69 | 0.92 | 0.87 | 0.00 | CBLN2 : cerebellum-specific protein involved in various signaling pathways. Possibly associated with pulmonary arterial hypertension. |
| NETO1 | 390,482 | NETO1 : brain-specific transmembrane protein involved in the regulation of neuronal circuitry. Associated with thyroid function. | |||||||
| All | SLC2A12 | 134,391,056 | rs4896021 | 17,267 | 0.76 | 0.96 | 0.95 | -0.01 | Catalyzes sugar absorption. Involved in the pathogenesis of diabetes. Associated with serum urate levels. |
| All | LOC101927657 | 127,358,509 | rs145200081 | 16,731 | 0.65 | 0.94 | 0.80 | 0.13 | Unknown (ncRNA). |
| All | LOC102724612 | 64,466,486 | rs113341339 | 78,446 | 0.73 | 0.91 | 0.95 | 0.00 | Unknown (ncRNA). |
| NE | ZBTB20 | 114,530,679 | rs9289004 | 10,658 | 0.55 | 0.65 | 0.82 | 0.07 | Transcriptional repressor associated with Primrose syndrome. |
| NE | ANXA10 | 168,646,016 | rs2176513 | 367,671 | 0.49 | 0.61 | 0.61 | -0.01 | Calcium-dependent phospholipid-binding annexin. |
| NE | TRPC3 | 122,905,041 | rs4502701 | 32,132 | 0.50 | 0.59 | 0.64 | -0.01 | Non-selective cation channel, associated with spinocerebellar ataxia. |
| NE | HS3ST1 | 11,634,592 | rs7665516 | 204,055 | 0.45 | 0.45 | 0.71 | 0.07 | Regulates rate of generation of anticoagulant heparan sulfate proteoglycan. |
| NE | MIR548C | 65,027,511 | rs2620721 | 11,126 | 0.50 | 0.55 | 0.73 | 0.03 | Unknown (microRNA). |
| NE | STARD13 | 33,799,901 | rs7318080 | 19,714 | 0.49 | 0.54 | 0.83 | 0.20 | Involved in cell proliferation and fibroblast morphology. |
| NE | AKAP11 | 42,931,386 | rs7319267 | 33,983 | 0.53 | 0.56 | 0.85 | 0.13 | Directs protein kinase A activity and is involved in cAMP messenger signaling. |
| NE | AGMO | 15,212,231 | rs35557899 | 27,711 | 0.47 | 0.51 | 0.68 | 0.01 | Catalyzes the cleavage of O-alkyl bonds of ether lipids. |
| NE | RUNX1T1 | 92,925,296 | rs11776341 | 41,898 | 0.45 | 0.56 | 0.54 | 0.00 | Involved in transcriptional repression. A translocation involving this gene is associated with acute myeloid leukemia. |
| NE | FHAD1 | 15,680,451 | rs2473358 | 971 | 0.45 | 0.60 | 0.52 | 0.00 | Unknown. |
| SW | KCNJ2 | 68,190,552 | rs35167900 | 14,369 | 0.57 | 0.61 | 0.93 | 0.22 | Potassium channel, associated with familial atrial fibrillation and periodic paralysis. |
| SW | TACC2 | 123,754,065 | rs10159998 | 5,062 | 0.50 | 0.60 | 0.67 | 0.00 | Belongs to a family of proteins that interact with the centrosome and microtubules, and that are implicated in cancer. |
| SW | LOC101928708 | 87,228,164 | rs4843556 | 17,556 | 0.58 | 0.65 | 0.86 | 0.07 | Unknown (ncRNA). |
| SW | C16orf82 | 27,187,689 | rs72782349 | 107,202 | 0.51 | 0.60 | 0.69 | 0.02 | Unknown. |
| SW | LOC100507391 | 194,520,805 | rs56379930 | 17,908 | 0.55 | 0.66 | 0.75 | -0.01 | Unknown (ncRNA). |
| SW | HAUS4 | 23,416,252 | rs2008951 | 127 | 0.49 | 0.50 | 0.83 | 0.16 | A component of a microtubule-binding complex that plays a role in the generation of microtubules in the mitotic spindle. |
| SW | KNG1 | 186,438,819 | rs5029990 | 815 | 0.51 | 0.56 | 0.72 | 0.01 | During the inflammatory response, it is involved in vasodilation, coagulation, enhanced capillary permeability and pain induction. |
| SW | MYDGF | 4,657,016 | rs66891175 | 540 | 0.55 | 0.61 | 0.88 | 0.16 | Unknown. |
| SW | MSMP | 35,757,075 | rs1951432 | 2,801 | 0.48 | 0.47 | 0.88 | 0.27 | May be involved in the tumorigenesis of prostate cancer. |
| SW | VAV2 | 136,756,316 | rs2519771 | 29,762 | 0.47 | 0.51 | 0.73 | 0.07 | Member of an oncogene family. Involved in T-cell receptor signaling. |
Top 10 peaks of differentiation from genome scans of all Aboriginal Australians combined (All) and two Aboriginal Australians subgroups living in different ecological regions in Australia.
RefSeq protein coding gene with exon boundary near to windowed-PBSn1 peak.
FST statistics at top SNP for each comparison within the PBSn1 calculation.
Genomic position (hg19) of SNP with highest value of PBSn1 within 200 Mb of the top window.
Distance between SNP and the nearest exon boundary of nearest gene.
PBSn1 statistic for top SNP.
Supplementary Material
References
- 1.Davidson I. The colonization of Australia and its adjacent islands and the evolution of modern cognition. Curr Anthropol. 2010;51:S177–S189. [Google Scholar]
- 2.Clarkson C, et al. The archaeology, chronology and stratigraphy of Madjedbebe (Malakunanja II): A site in northern Australia with early occupation. J Hum Evol. 2015;83:46–64. doi: 10.1016/j.jhevol.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 3.O’Connell JF, Allen J. The process, biotic impact, and global implications of the human colonization of Sahul about 47,000 years ago. J Archaeol Sci. 2015;56:73–84. [Google Scholar]
- 4.Barker G, et al. The ‘human revolution’in tropical Southeast Asia: the antiquity of anatomically modern humans, and of behavioural modernity, at Niah Cave (Sarawak, Borneo) J Hum Evol. 2007;52:243–261. doi: 10.1016/j.jhevol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Lahr MM, Foley R. Multiple dispersals and modern human origins. Evol Anthropol Issues News Rev. 1994;3:48–60. [Google Scholar]
- 6.Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press; 1996. [Google Scholar]
- 7.Wollstein A, et al. Demographic History of Oceania Inferred from Genome-wide Data. Curr Biol. 2010;20:1983–1992. doi: 10.1016/j.cub.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen M, et al. An Aboriginal Australian Genome Reveals Separate Human Dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reich D, et al. Denisova Admixture and the First Modern Human Dispersals into Southeast Asia and Oceania. Am J Hum Genet. 2011;89:516–528. doi: 10.1016/j.ajhg.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves JM, et al. Climate variability over the last 35,000 years recorded in marine and terrestrial archives in the Australian region: an OZ-INTIMATE compilation. Quat Sci Rev. 2013;74:21–34. [Google Scholar]
- 11.Hiscock P, Wallis LA. In: Desert Peoples. Veth P, Smith M, Hiscock P, editors. Blackwell Publishing Ltd; 2005. pp. 34–57. [Google Scholar]
- 12.Birdsell JB. Microevolutionary Patterns in Aboriginal Australia: A Gradient Analysis of Clines. Oxford University Press; 1993. [Google Scholar]
- 13.Bowern C, Atkinson Q. Computational phylogenetics and the internal structure of Pama-Nyungan. Language. 2012;88:817–845. [Google Scholar]
- 14.Dixon RMW. Australian Languages: Their Nature and Development. Cambridge University Press; 2002. [Google Scholar]
- 15.Evans N, McConvell P. The enigma of Pama-Nyungan expansion in Australia. Archaeol Lang II. 1997:174–191. [Google Scholar]
- 16.Hiscock Peter. Archaeology of ancient Australia. Routledge; 2008. [Google Scholar]
- 17.Bellwood P. First Migrants: Ancient Migration in Global Perspective. Wiley-Blackwell; 2013. [Google Scholar]
- 18.Pugach I, Delfin F, Gunnarsdóttir E, Kayser M, Stoneking M. Genome-wide data substantiate Holocene gene flow from India to Australia. Proc Natl Acad Sci U S A. 2013;110:1803–1808. doi: 10.1073/pnas.1211927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macknight CC. Macassans and the Aboriginal Past. Archaeol Ocean. 1986;21:69–75. [Google Scholar]
- 20.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frichot E, Mathieu F, Trouillon T, Bouchard G, François O. Fast and Efficient Estimation of Individual Ancestry Coefficients. Genetics. 2014;196:973–983. doi: 10.1534/genetics.113.160572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson NJ, et al. Ancient Admixture in Human History. Genetics genetics. 2012 doi: 10.1534/genetics.112.145037. 112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Excoffier L, Dupanloup I, Huerta-Sanchez E, Sousa VC, Foll M. Robust Demographic Inference from Genomic and SNP Data. PLoS Genet. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Birdsell JB. Preliminary data on the trihybrid origin of the Australian Aborigines. Archaeol Phys Anthropol Ocean. 1967:100–155. [Google Scholar]
- 25.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin P, Stoneking M. Denisovan Ancestry in East Eurasian and Native American Populations. Mol Biol Evol. 2015 doi: 10.1093/molbev/msv141. msv141. [DOI] [PubMed] [Google Scholar]
- 27.Fu Q, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the Joint Demographic History of Multiple Populations from Multidimensional SNP Frequency Data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bergström A, et al. Deep Roots for Aboriginal Australian Y Chromosomes. Curr Biol. 2016 doi: 10.1016/j.cub.2016.01.028. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudjashov G, et al. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc Natl Acad Sci. 2007;104:8726–8730. doi: 10.1073/pnas.0702928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lippold S, et al. Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences. Investig Genet. 2014;5:13. doi: 10.1186/2041-2223-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radcliffe-Brown AR. The Social Organization of Australian Tribes. Oceania. 1930;1:34–63. [Google Scholar]
- 33.Clark PU, et al. The last glacial maximum. Science. 2009;325:710–714. doi: 10.1126/science.1172873. [DOI] [PubMed] [Google Scholar]
- 34.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian Coalescent Inference of Past Population Dynamics from Molecular Sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 35.Lourandos H, David B. In: in Bridging Wallace’s Line: the Environmental and Cultural History and Dynamics of the SE Asian-Australasian Region. Kershaw AP, David B, Tapper N, Penny D, Brown J, editors. pp. 97–118. [Google Scholar]
- 36.Evans N, Jones R. Archaeology and linguistics: aboriginal Australia in global perspective. Oxford University Press Australia; 1997. [Google Scholar]
- 37.Yi X, et al. Sequencing of 50 Human Exomes Reveals Adaptation to High Altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qi X, Chan WL, Read RJ, Zhou A, Carrell RW. Temperature-responsive release of thyroxine and its environmental adaptation in Australians. Proc R Soc Lond B Biol Sci. 2014;281 doi: 10.1098/rspb.2013.2747. 20132747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tin A, et al. Genome-wide association study for serum urate concentrations and gout among African Americans identifies genomic risk loci and a novel URAT1 loss-of-function allele. Hum Mol Genet. 2011;20:4056–4068. doi: 10.1093/hmg/ddr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 41.Fenner JN. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am J Phys Anthropol. 2005;128:415–423. doi: 10.1002/ajpa.20188. [DOI] [PubMed] [Google Scholar]
- 42.Holt S. Palaeoenvironments of the Gulf of Carpentaria from the last glacial maximum to the present, as determined by foraminiferal assemblages. 2005 [Google Scholar]
- 43.Horton D. The Encyclopedia of Aboriginal Australia. Australian Institute of Aboriginal and torres Strait Islander Studies; 1994. [Google Scholar]
- 44.Migliano A, et al. Evolution of the Pygmy Phenotype: Evidence of Positive Selection from Genome-wide Scans in African, Asian, and Melanesian Pygmies. Hum Biol. 2013;85 doi: 10.3378/027.085.0313. [DOI] [PubMed] [Google Scholar]
- 45.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wall JD, et al. Higher Levels of Neanderthal Ancestry in East Asians than in Europeans. Genetics. 2013;194:199–209. doi: 10.1534/genetics.112.148213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vernot B, Akey JM. Resurrecting Surviving Neandertal Lineages from Modern Human Genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 48.Fu Q, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, et al. Comparing spatial maps of human population-genetic variation using Procrustes analysis. Stat Appl Genet Mol Biol. 2010;9 doi: 10.2202/1544-6115.1493. Article 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.