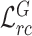Abstract
The novel Coronavirus disease (COVID-19) is a highly contagious virus and has spread all over the world, posing an extremely serious threat to all countries. Automatic lung infection segmentation from computed tomography (CT) plays an important role in the quantitative analysis of COVID-19. However, the major challenge lies in the inadequacy of annotated COVID-19 datasets. Currently, there are several public non-COVID lung lesion segmentation datasets, providing the potential for generalizing useful information to the related COVID-19 segmentation task. In this paper, we propose a novel relation-driven collaborative learning model to exploit shared knowledge from non-COVID lesions for annotation-efficient COVID-19 CT lung infection segmentation. The model consists of a general encoder to capture general lung lesion features based on multiple non-COVID lesions, and a target encoder to focus on task-specific features based on COVID-19 infections. We develop a collaborative learning scheme to regularize feature-level relation consistency of given input and encourage the model to learn more general and discriminative representation of COVID-19 infections. Extensive experiments demonstrate that trained with limited COVID-19 data, exploiting shared knowledge from non-COVID lesions can further improve state-of-the-art performance with up to 3.0% in dice similarity coefficient and 4.2% in normalized surface dice. In addition, experimental results on large scale 2D dataset with CT slices show that our method significantly outperforms cutting-edge segmentation methods metrics. Our method promotes new insights into annotation-efficient deep learning and illustrates strong potential for real-world applications in the global fight against COVID-19 in the absence of sufficient high-quality annotations.
Keywords: COVID-19, computed tomography, few-shot learning, knowledge transfer, lung infection segmentation
I. Introduction
Since the beginning of 2020, the novel coronavirus disease (COVID-19) has rapidly spread worldwide, posing an extremely serious threat and challenge to all countries. This severe disease has been declared as a public health emergency of international concern by the World Health Organization (WHO), which has caused more than 2,600,000 deaths until the date of 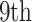 March 2021, according to the statistics of Johns Hopkins Coronavirus Resource Center.1
March 2021, according to the statistics of Johns Hopkins Coronavirus Resource Center.1
As one of the most commonly used imaging methods, computed tomography (CT) plays an important role in the fight against COVID-19 [1]–[3]. Researchers have proved that CT images have strong ability to capture typical features like ground glass and bilateral patchy shadows of affected patients [4] and are shown to be more sensitive compared with standard viral nucleic acid detection using real-time polymerase chain reaction (RT-PCR) for the early diagnosis of COIVD-19 infection [5]. Besides, CT images can provide visual evaluation of the extent of lung abnormalities and assist the process of prognostic [6].
In clinical practice, the segmentation of lung infections from CT images is an important component to assist in further assessment and quantification of the diseases [7]. Since manual contour delineation is time-consuming and laborious, and suffers from inter and intra-observer variabilities [8], it is of great significance to develop artificial intelligence-based approaches to assist in the automatic segmentation of COVID-19 infections. Recently, the unprecedented development in deep learning has showed significant improvements and achieved state-of-the-art performances in many medical image segmentation tasks [9]–[12], and deep neural networks have been widely applied in the global fight against COVID-19 [13]–[16].
However, the success of deep learning methods mainly requires large amount of high-quality annotated datasets, while it is impractical to collect large amount of well annotated data in real clinical approach, especially when radiologists are busy fighting the coronavirus disease. Additionally, as shown in Fig. 1, the large variations in shape, size and position of lung infections and large inter-case variations pose great challenges for the segmentation tasks [17]. Therefore, exploring annotation-efficient COVID-19 lung infection segmentation methods with limited labeled data has become an urgent need especially in the current situation.
Fig. 1.
Examples of COVID-19 infections in CT volumes showing the large variations of shape, size and position of lung infections. The upper row shows the raw images (from axial view) and the lower row shows corresponding annotations of infection areas.
Currently, there are several public non-COVID lung lesion datasets due to other clinical practices, such as MSD Lung for segmentation of lung tumor and NSCLC Pleural Effusion for segmentation of pleural effusion. These non-COVID datasets may serve as potential profit for generalizing useful information to the related COVID-19 infection segmentation task. Wang et al. [18] have proven that pre-training on non-COVID datasets can improve the segmentation performance of COVID-19 infection segmentation. However, the improvement of transfer learning is not stable when encountering large domain difference between datasets, and shared knowledge between COVID-19 and non-COVID lung lesions cannot be fully exploited.
To address these challenges, we propose a novel relation-driven collaborative learning model for annotation-efficient COVID-19 CT lung infection segmentation by exploiting shared knowledge from non-COVID lesions. The network consists of encoders with the same architecture and a shared decoder. The general encoder is adopted to capture general lung lesion features based on multiple non-COVID lesions, while the target encoder is adopted to focus on task-specific features of COVID-19 infections. Features extracted from the two parallel encoders are concatenated for the subsequent decoder part. To exploit shared knowledge between COVID and non-COVID lesions, we develop a collaborative learning scheme to regularize the relation consistency between extracted features of given input. Our method can enforce the consistency of feature relation among extracted features and encourage the model to explore semantic information from both COVID-19 and non-COVID cases. Besides, the scheme can also be extended to utilize unlabeled COVID-19 data for feature relation regularization and achieve more consistent and robust learning. The contributions of this work are summarized as follows:
-
•
We propose a novel relation-driven collaborative learning model for annotation-efficient segmentation of COVID-19 lung infections from CT images by leveraging shared knowledge from non-COVID lesions to improve the segmentation performance of COVID-19 infections with limited training data.
-
•
We present a collaborative learning scheme to explore general semantic information from both COVID-19 and non-COVID cases by regularizing feature-level relation consistency of given input, so as to encourage the model to learn more general and discriminative representation of COVID-19 infections for better segmentation performance. The scheme can also be extended to utilize unlabeled COVID-19 data for the regularization to achieve more consistent and robust learning.
-
•
We have conducted extensive experiments on two COVID-19 datasets and two non-COVID lung lesion datasets for 2D and 3D segmentation tasks. The results show that our method achieves superior segmentation performance compared with other methods in the absence of sufficient high-quality COVID-19 data.
II. Related Work
In this section, we briefly review the research related to our work. We first review works on annotation-efficient deep learning for medical image segmentation. Then we review existing works on COVID-19 segmentation and transfer learning approaches for COVID-19.
A. Annotation-Efficient Deep Learning
Compared with natural images, the annotations of medical images are much harder and more expensive to acquire due to following problems: 1) annotating medical images heavily relies on professional diagnosis knowledge of radiologists; 2) most modalities of medical images like CT are 3D volumes, which will take much more time and labor for annotation. To alleviate annotation scarcity, annotation-efficient methods have received great attention in medical image analysis community [19], [20]. For example, semi-supervised learning aims at learning from a limited amount of labeled data and a large amount of unlabeled data, which is an effective way to explore knowledge from the unlabeled data [21]. Weakly supervised learning explores the use of weak annotations like noisy annotations and sparse annotations [22]. Besides, some approaches also aim at integrating multiple related datasets to learn general knowledge [23], [24]. To issue the problem of limited labeled COVID-19 data, in this work, we aim at utilizing existing non-COVID lung lesion datasets for generalizing useful information to related COVID-19 task, so as to achieve better segmentation performance with limited in-domain training data.
B. Research on COVID-19 Segmentation
Automatic segmentation of COVID-19 infections from CT images is a crucial step to for quantification of the disease progression. Recently, several approaches have been proposed for COVID-19 lung infection segmentation. Shan et al. [25] propose a deep learning-based system for automatic segmentation and quantification of infection regions. Amyar et al. [26] propose to improve the segmentation performance with a multi-task learning approach. Xie et al. [27] propose a relational approach to leverages structured relationships by introducing a novel non-local neural network module to learn both visual and geometric relationships. Zhou et al. [28] propose a U-Net based segmentation network to incorporate spatial and channel attention for better feature representation. Other than fully supervised learning, Zheng et al. [29] develop a weakly-supervised approach to investigate the potential for automatic detection of COVID-19 based on patient-level label. Fan et al. [30] present a lung infection segmentation network for 2D CT slices with semi-supervised strategy. Wang et al. [31] propose a noise-robust framework to learn from noisy labels for the pneumonia lesion segmentation task. Yao et al. [32] use a set of operations to synthesize lesion-like appearances for label-free segmentation. Ma et al. [33] propose an active contour regularized framework using region-scalable fitting to regularize and refine the pseudo labels for semi-supervised infection segmentation.
C. Transfer Learning Approaches for COVID-19
Transfer learning aims to leverage knowledge and latent features from other datasets by pre-training models on large datasets and fine-tuning trained models on downstream tasks. Due to the problem of limited COVID-19 data, several transfer learning methods have been applied. For example, Chouhan et al. [34] propose an ensemble model to combine outputs from five pre-trained models based on ImageNet. Majeed et al. [35] adopt transfer learning procedure and propose a simple CNN architecture with a small number of parameters to distinguish COVID-19 from normal X-rays. Misra et al. [36] propose a multi-channel pre-trained ResNet architecture to facilitate the diagnosis of COVID-19. For segmentation of COVID-19 infections, Wang et al. [18] evaluate different transfer learning methods and revealed the benefits of transferring knowledge from non-COVID lung lesions. However, transfer learning only takes the advantage of existing models, and non-COVID cases are not utilized in the training procedure of downstream COVID-19 segmentation tasks. Different from these existing methods, our method aims at learning from COVID-19 and non-COVID lung lesions collaboratively to exploit shared semantic information.
III. Method
In this section, we first introduce the overview of our proposed method. Then we provide details of our relation-driven collaborative learning scheme and the overall training procedure.
A. Overview
An overview of our proposed framework is shown in Fig. 2. Following the design of standard U-Net [37], [38], our network consists of two encoders with the same architecture and a shared decoder. Since the encoder serves as a contraction to extract image contextual features, the upper one named general encoder (G) is adopted to capture general lung lesion features based on multiple non-COVID lesions, and the lower one named target encoder (T) is adopted to focus on task-specific features of the target COVID-19 infection segmentation task. After that, extracted features from these two parallel encoders are concatenated together for the input of the decoder. The shared decoder (D) serves as a symmetric expanding path to recover the spatial information of the extracted features. Since our motivation is to use non-COVID lesions to assist in the segmentation of COVID-19 infections, the general encoder can be seemed as an auxiliary branch to extract shared knowledge. Therefore, we only employ the skip connections between the target encoder and shared decoder for the fusion of multi-scale features as shown in the top right corner of Fig. 2.
Fig. 2.

The overview of our proposed relation-driven collaborative learning model, where green and blue represent the data flow of general encoder and target encoder for COVID-19 infection segmentation, respectively. Extracted features from these two parallel encoders are concatenated for the input of the shared decoder. To exploit shared knowledge from non-COVID-cases, an additional data flow in orange is adopted. By regularizing feature-level relation consistency of given input, the model is encouraged to explore semantic information from both COVID-19 and non-COVID cases. Since the general encoder is applied to utilize non-COVID data and assist in the learning of COVID-19 segmentation as an auxiliary branch, we only employ the skip connections between the target encoder and shared decoder for the fusion of multi-scale features as shown in the top right corner.
Given a set of samples 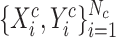 from COVID-19 datasets
from COVID-19 datasets 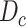 and a set of samples
and a set of samples 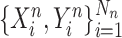 from non-COVID datasets
from non-COVID datasets 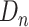 , where
, where 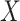 and
and 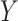 denote the CT image and corresponding annotation of lung lesions. For the segmentation workflow, the general encoder is applied to extract general features, while the target encoder is applied to extract the task-specific features. These extracted features are concatenated and then fed into the decoder part to get the final segmentation results. To issue the problem of limited COVID-19 training data, instead of transferring pre-trained models to the downstream learning task of
denote the CT image and corresponding annotation of lung lesions. For the segmentation workflow, the general encoder is applied to extract general features, while the target encoder is applied to extract the task-specific features. These extracted features are concatenated and then fed into the decoder part to get the final segmentation results. To issue the problem of limited COVID-19 training data, instead of transferring pre-trained models to the downstream learning task of  , we aim to involve
, we aim to involve 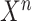 collaboratively in the training procedure of COVID-19 to exploit shared knowledge from non-COVID cases, which can be used as a guidance for the learning of target COVID-19 infection segmentation. Specifically, a relation-driven collaborative learning scheme is applied to regularize the relation consistency between extracted features of given input and encourage the model to explore semantic information.
collaboratively in the training procedure of COVID-19 to exploit shared knowledge from non-COVID cases, which can be used as a guidance for the learning of target COVID-19 infection segmentation. Specifically, a relation-driven collaborative learning scheme is applied to regularize the relation consistency between extracted features of given input and encourage the model to explore semantic information.
B. Relation-Driven Collaborative Learning
Inspired by recent study on data-level regularization with sample relation [39], to exploit shared knowledge between non-COVID and COVID-19 cases for collaborative learning, we propose to regularize feature-level relation consistency of given input, so as to facilitate the learning procedure of COVID-19 lung infections segmentation. Based on the assumption that general encoder is adopted to capture general features of lung lesions, and the target encoder is adopted to focus on task-specific features of COVID-19 infection, we aim to enforce the relation of features extracted from these two encoders as guidance for the collaborative learning approach.
To estimate the relation of extracted features, we model the feature relation with channel-wise Gram Matrix [40]. For each input batch with 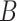 samples, we average the features within each batch to get the mean representation. We denote the extracted feature maps of encoder as
samples, we average the features within each batch to get the mean representation. We denote the extracted feature maps of encoder as 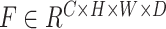 for 3D segmentation networks or
for 3D segmentation networks or 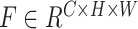 for 2D segmentation networks, where
for 2D segmentation networks, where  ,
, 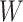 and
and 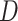 represent the spatial dimension of feature maps, and
represent the spatial dimension of feature maps, and 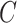 represents the channel number. To obtain channel-wise feature relation, we reshape the feature maps into
represents the channel number. To obtain channel-wise feature relation, we reshape the feature maps into  or
or 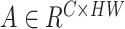 . After that, we get the channel-wise Gram Matrix as follows:
. After that, we get the channel-wise Gram Matrix as follows:
 |
where the value of  row and
row and 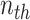 column
column 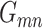 is the inner product between the vectorized activation maps
is the inner product between the vectorized activation maps  and
and 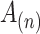 , representing the similarity between the
, representing the similarity between the 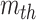 and
and 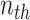 channel of extracted features. Therefore, the final feature relation matrix
channel of extracted features. Therefore, the final feature relation matrix  is obtained by conducting the
is obtained by conducting the 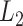 normalization for each row of
normalization for each row of 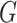 as follows
as follows
 |
After the modeling of feature relation, our method encourages the network to learn more general and discriminative representation of COVID-19 infections by regularizing the feature relation consistency among given input, so as to explore semantic information from both COVID-19 and non-COVID cases.
For explicit learning, the network is optimized based on the supervised segmentation loss 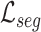 between output
between output  and corresponding ground truth
and corresponding ground truth 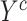 . We use the combination of dice loss
. We use the combination of dice loss 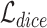 and cross entropy loss
and cross entropy loss 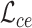 as the supervised segmentation loss, and deep supervision [41] is applied to obtain multi-scale supervision at different scales. The supervised segmentation loss can be summarized as
as the supervised segmentation loss, and deep supervision [41] is applied to obtain multi-scale supervision at different scales. The supervised segmentation loss can be summarized as
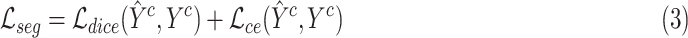 |
Besides, to utilize the feature relation for collaborative learning, the non-COVID cases are additionally fed into general encoder to explore the general feature representation and its corresponding feature relation matrix  . To ensure that general encoder can capture general features of lung lesions, our proposed scheme requires the generated feature relation matrices of general encoder to be stable using general relation consistency loss
. To ensure that general encoder can capture general features of lung lesions, our proposed scheme requires the generated feature relation matrices of general encoder to be stable using general relation consistency loss  defined as
defined as
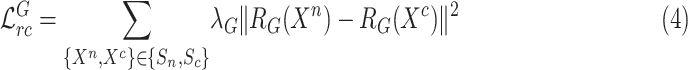 |
While for target encoder, we enforce the extracted relation matrices of task-specific features to be more discriminative compared with general encoder using target relation consistency loss 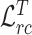 defined as
defined as
 |
where 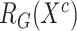 and
and 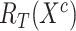 denote the feature relation matrices of COVID-19 cases extracted from general encoder and target encoder, respectively.
denote the feature relation matrices of COVID-19 cases extracted from general encoder and target encoder, respectively. 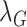 and
and  are ramp-up weighting coefficients that control the trade-off between the segmentation loss and consistency loss, so as to mitigate the disturbance of consistency loss at early training stage. Since the network is supervised by limited COVID-19 cases, the training may become unstable and with poor generalization ability. By minimizing feature relation consistency losses
are ramp-up weighting coefficients that control the trade-off between the segmentation loss and consistency loss, so as to mitigate the disturbance of consistency loss at early training stage. Since the network is supervised by limited COVID-19 cases, the training may become unstable and with poor generalization ability. By minimizing feature relation consistency losses 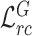 and
and 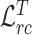 during the training procedure, the general encoder and target encoder can be enhanced to capture more general and discriminative representation, thereby exploring useful shared knowledge from adequate non-COVID data for better segmentation performance.
during the training procedure, the general encoder and target encoder can be enhanced to capture more general and discriminative representation, thereby exploring useful shared knowledge from adequate non-COVID data for better segmentation performance.
Algorithm 1: Training Procedure of Our Proposed Framework.
Input:A batch of (
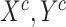 ) from COVID-19 dataset
) from COVID-19 dataset 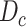 and (
and ( ) from non-COVID dataset
) from non-COVID dataset  .
.Output:Trained network
 with parameters
with parameters 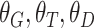
-
1:
while not converge do
-
2:
(
 ), (
), (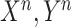 )
) 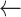 sampled from
sampled from 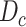 and
and 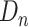
-
3:
Generate features of general encoder
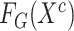 and target encoder
and target encoder 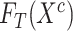
-
4:
Generate general feature representation
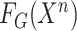
-
5:
Calculate feature relation matrices
 ,
, 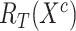 and
and 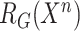 as Eq. (1) and (2)
as Eq. (1) and (2) -
6:
Generate segmentation output

-
7:
Calculate segmentation loss
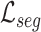 as Eq. (3)
as Eq. (3) - 8:
-
9:
Update
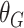

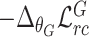
-
10:
Update

-
11:
Update


-
12:
Ramp up the weighting coefficients
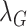 and
and 
-
13:
end while
-
14:
returnTrained network

C. Overall Training Procedure
Algorithm1 presents the detailed training procedure of our framework. For the optimation of the network, we update the target encoder and decoder based on the supervised segmentation loss 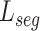 . Besides, relation consistency losses
. Besides, relation consistency losses  and
and 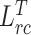 are used to update the general encoder and target encoder, respectively. The collaborative learning scheme allow the two parallel encoders to benefit from each other's guidance, encouraging the model to explore semantic information from both COVID-19 and non-COVID cases.
are used to update the general encoder and target encoder, respectively. The collaborative learning scheme allow the two parallel encoders to benefit from each other's guidance, encouraging the model to explore semantic information from both COVID-19 and non-COVID cases.
D. Feature Relation Regularization With Unlabeled Data
Since our proposed general relation consistency loss  and target relation consistency loss
and target relation consistency loss  do not require segmentation label, our proposed method can be straightforwardly extended to utilize unlabeled COVID-19 data for feature relation regularization. Specifically, we only activate the supervised segmentation loss
do not require segmentation label, our proposed method can be straightforwardly extended to utilize unlabeled COVID-19 data for feature relation regularization. Specifically, we only activate the supervised segmentation loss 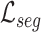 for labeled data, while computing the relation consistency losses
for labeled data, while computing the relation consistency losses 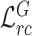 and
and  for all the training data. In this way, unlabeled data can be leveraged for the regularization to achieve more consistent and robust learning.
for all the training data. In this way, unlabeled data can be leveraged for the regularization to achieve more consistent and robust learning.
IV. Experiments
A. Dataset Introduction
1). COVID-19 Dataset
We select out two public COVID-19 lung infection segmentation datasets for our experiments. The first dataset contains 20 CT volumes with over 1800 annotated slices released by Coronacases Initiative and Radiopaedial, which is publicly available at.2 The annotation of infections is labeled by two radiologists and verified by an experienced radiologist by Ma et al. [42]. The second dataset is COVID-19 CT Segmentation dataset3 collected by the Italian Society of Medical and Interventional Radiology, which contains 100 2D axial CT slices from different COVID-19 patients. A radiologist segmented the CT images using different labels for identifying lung infections.
2). Non-COVID Lung Lesion Datasets
In order to explore relevant information from non-COVID lung lesions to promote the annotation-efficient training of COVID-19 cases, we select out two public non-COVID lung lesion segmentation datasets for our following experiments. The first dataset is MSD Lung Tumor Dataset of Medical Segmentation Decathlon (MSD) Challenge [43] in MICCAI 2018.4 This dataset is comprised of patients with non-small cell lung cancer from Stanford University (Palo Alto, CA, USA) publicly available through TCIA. The tumor is annotated by an expert thoracic radiologist and 63 labeled CT volumes are used. The second dataset is NSCLC Pleural Effusion Dataset.5 This dataset contains 78 CT volumes with annotation of pleural effusion. To exploit general features of lung lesions, we combine MSD and NSCLC datasets to form a non-COVID multi-lesion dataset in the following experiments.
B. Experimental Settings
1). 3D Experiments on CT Volumes
For 3D experiments of CT Volumes, to make a fair comparison, we follow the task settings of COVID-19 benchmarks in [42]. For the COVID-19 dataset, we make the same 5-fold cross validation based on pre-defined dataset split. Each fold contains 4 scans (20%) for training and 16 scans (80%) for testing. For non-COVID lung lesion datasets, we randomly select 80% of the data for training and the rest of 20% for validation.
We use 3D U-Net [38] as the backbone network. Details of network architecture is shown in Table I. The input patch size is set as 56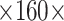 192 with batch size of 2. Stochastic gradient descent (SGD) optimizer is used for training with initial learning rate of 0.01 and momentum of 0.99.
192 with batch size of 2. Stochastic gradient descent (SGD) optimizer is used for training with initial learning rate of 0.01 and momentum of 0.99.
TABLE I. Details of 3D U-Net Architecture Used in Our Experiments.
| feature size | Encoder (G / T) | Decoder (D) | ||
|---|---|---|---|---|
| 1x56x160x192 | input | output | conv(1x1x1)-sigmoid | |
| 32x56x160x192 | conv1 | conv(1x3x3)-IN-LReLU | conv10 | conv(1x3x3)-IN-LReLU |
| 64x56x80x96 | down1 | strided conv(1,2,2) | up10 | transposed conv(1,2,2) - conv1 |
| 64x56x80x96 | conv2 | conv(3x3x3)-IN-LReLU | conv9 | conv(3x3x3)-IN-LReLU |
| 128x28x40x48 | down2 | strided conv(2,2,2) | up9 | transposed conv(2,2,2) - conv2 |
| 128x28x40x48 | conv3 | conv(3x3x3)-IN-LReLU | conv8 | conv(3x3x3)-IN-LReLU |
| 256x14x20x24 | down3 | strided conv(2,2,2) | up8 | transposed conv(2,2,2) - conv3 |
| 256x14x20x24 | conv4 | conv(3x3x3)-IN-LReLU | conv7 | conv(3x3x3)-IN-LReLU |
| 320x7x10x12 | down4 | strided conv(2,2,2) | up7 | transposed conv(2,2,2) - conv4 |
| 320x7x10x12 | conv5 | conv(3x3x3)-IN-LReLU | conv6 | conv(3x3x3)-IN-LReLU |
| 320x7x5x6 | down5 | strided conv(1,2,2) | up6 | transposed conv(1,2,2) - conv5 |
Note that the general encoder and target encoder are with the same architecture as shown in the left column.
2). 2D Experiments on Axial CT Slices
To compare our method with state-of-the-art methods for 2D medical image segmentation, we make comparison experiments based on 2D COVID-19 CT slices. Following the same task settings of [30], we use the same 50 images for training and validation, and the remaining 50 slices for testing. For non-COVID lung lesion datasets, we randomly select out 100 2D slices with lung lesions from different CT scans. Besides, unlabeled training set of COVID-SemiSeg Dataset [30] is used to evaluate the effectiveness of our proposed method to utilize unlabeled COVID-19 data.
We use 2D U-Net [37] as the backbone network. Details of network architecture is shown in Table II. The input patch size is set as 448×384 with batch size of 2. Stochastic gradient descent (SGD) optimizer is used for training with initial learning rate of 0.01 and momentum of 0.99.
TABLE II. Details of 2D U-Net Architecture Used in Our Experiments.
| feature size | Encoder (G / T) | Decoder (D) | ||
|---|---|---|---|---|
| 1x448x384 | input | output | conv(1x1)-sigmoid | |
| 32x448x384 | conv1 | conv(3x3)-IN-LReLU | conv12 | conv(3x3)-IN-LReLU |
| 64x224x192 | down1 | strided conv(2,2) | up12 | transposed conv(2,2) - conv1 |
| 64x224x192 | conv2 | conv(3x3)-IN-LReLU | conv11 | conv(3x3)-IN-LReLU |
| 128x112x96 | down2 | strided conv(2,2) | up11 | transposed conv(2,2) - conv2 |
| 128x112x96 | conv3 | conv(3x3)-IN-LReLU | conv10 | conv(3x3)-IN-LReLU |
| 256x56x48 | down3 | strided conv(2,2) | up10 | transposed conv(2,2) - conv3 |
| 256x56x48 | conv4 | conv(3x3)-IN-LReLU | conv9 | conv(3x3)-IN-LReLU |
| 480x28x24 | down4 | strided conv(2,2) | up9 | transposed conv(2,2) - conv4 |
| 480x28x24 | conv5 | conv(3x3)-IN-LReLU | conv8 | conv(3x3)-IN-LReLU |
| 480x14x12 | down5 | strided conv(2,2) | up8 | transposed conv(2,2) - conv5 |
| 480x14x12 | conv6 | conv(3x3)-IN-LReLU | conv7 | conv(3x3)-IN-LReLU |
| 480x7x6 | down6 | strided conv(2,2) | up7 | transposed conv(2,2) - conv6 |
Note that the general encoder and target encoder are with the same architecture as shown in the left column.
C. Implementation Details and Evaluation Metrics
All the experiments in our work are implemented in Pytorch [44] and trained on NVIDIA Tesla V100 GPUs. Our backbone network is based on nnUNet [45] that achieved state-of-the-art results in 23 segmentation challenges with automatically designed U-Net according to the dataset properties. To unify the setting for our collaborative learning approach, we use planned network architectures of COVID-19 infection segmentation task for our framework. Following [46], we use a Gaussian ramp-up function 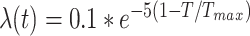 to control the balance between supervised loss and consistency loss, where
to control the balance between supervised loss and consistency loss, where 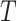 represents the current training step and
represents the current training step and  represents the maximum training step.
represents the maximum training step.
Motivated by the evaluation methods of the medical image segmentation decathlon [43], we employ two complementary metrics to evaluate the segmentation performance. Dice Similarity Coefficient (DSC), a region-based measure is used to measure the region mismatch, and Normalized surface Dice (NSD), a boundary-based measure is used to evaluate how close the segmentation and ground truth surfaces are to each other. Both metrics take the values in [0,1] and higher scores represent better segmentation performance. Let 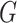 and
and 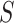 denote the ground truth and the segmentation result, respectively. The two metrics are defined as follows:
denote the ground truth and the segmentation result, respectively. The two metrics are defined as follows:
 |
where 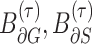 denote the border regions of ground truth and segmentation surface at a threshold
denote the border regions of ground truth and segmentation surface at a threshold 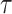 to tolerate the inter-rater variability of the annotators. We set
to tolerate the inter-rater variability of the annotators. We set  = 3 mm for the evaluation of segmentation results in the following experiments. Besides, we also consider three other evaluation metrics in 2D experiments. Sensitivity (Sen) denotes the percentage of positive instances correctly identified. Specificity (Spec) denotes the percentage of predicted positive instances that are correctly identified. Mean Absolute Error (MAE) measures the pixel-wise error between segmentation output and corresponding groundtruth.
= 3 mm for the evaluation of segmentation results in the following experiments. Besides, we also consider three other evaluation metrics in 2D experiments. Sensitivity (Sen) denotes the percentage of positive instances correctly identified. Specificity (Spec) denotes the percentage of predicted positive instances that are correctly identified. Mean Absolute Error (MAE) measures the pixel-wise error between segmentation output and corresponding groundtruth.
D. Ablation Analysis
To evaluate the effectiveness of the key components in our framework, we conduct ablation studies by removing the feature relation consistency loss. As shown in Table III, it is observed that all our methods can achieve better performance on all metrics compared with fully supervised methods, showing the effectiveness of our method. Besides, the usage of  and
and 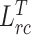 can both further improve the segmentation performance compared with baseline. When removing the target relation consistency, the average segmentation performance of five folds is degraded by 0.4% and 0.2% on DSC and NSD, respectively. The result proves that the usage of target relation consistency loss
can both further improve the segmentation performance compared with baseline. When removing the target relation consistency, the average segmentation performance of five folds is degraded by 0.4% and 0.2% on DSC and NSD, respectively. The result proves that the usage of target relation consistency loss  can enforce the target encoder to be more discriminative, so as to improve the segmentation performance. However, the improvement is susceptible to the domain difference. Besides, we also conduct experiments of our backbone by removing the general relation consistency loss
can enforce the target encoder to be more discriminative, so as to improve the segmentation performance. However, the improvement is susceptible to the domain difference. Besides, we also conduct experiments of our backbone by removing the general relation consistency loss 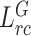 . In this way, the general encoder is frozen and are not updated during the training procedure, which means that the knowledge transfer is not available. The experimental results demonstrate that the average segmentation performance is degraded by 0.6% and 0.3% on DSC and NSD, showing the importance of knowledge transfer in our collaborative learning scheme. Some segmentation results of our method and 3D nnUNet are illustrated in Fig. 3 for visual comparison. As shown in the figure, our method can generate segmentation results with more accurate boundaries in Fig. 3(a)(b), and less segmentation mistakes in small infection areas in Fig. 3(c)(d)(e). These results demonstrate that the collaborative learning approach can better exploit shared knowledge from non-COVID cases, leading to better performance when generalizing on test data.
. In this way, the general encoder is frozen and are not updated during the training procedure, which means that the knowledge transfer is not available. The experimental results demonstrate that the average segmentation performance is degraded by 0.6% and 0.3% on DSC and NSD, showing the importance of knowledge transfer in our collaborative learning scheme. Some segmentation results of our method and 3D nnUNet are illustrated in Fig. 3 for visual comparison. As shown in the figure, our method can generate segmentation results with more accurate boundaries in Fig. 3(a)(b), and less segmentation mistakes in small infection areas in Fig. 3(c)(d)(e). These results demonstrate that the collaborative learning approach can better exploit shared knowledge from non-COVID cases, leading to better performance when generalizing on test data.
TABLE III. Quantitative Results of 5-Fold Cross Validation of Ablation Analysis in Our Experiments.
| Method | DSC | NSD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold 0 | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Avg | Fold 0 | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Avg | |
| 3D nnUNet | 0.681 | 0.713 | 0.662 | 0.681 | 0.627 | 0.673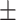 0.223 0.223 |
0.709 | 0.718 | 0.717 | 0.708 | 0.649 | 0.700 0.224 0.224 |
| Ours (baseline) | 0.689 | 0.721 | 0.712 | 0.720 | 0.632 | 0.695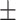 0.205 0.205 |
0.709 | 0.747 | 0.770 | 0.764 | 0.649 | 0.728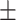 0.216 0.216 |
Ours (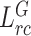 ) ) |
0.701 | 0.727 | 0.727 | 0.710 | 0.625 | 0.699 0.210 0.210 |
0.747 | 0.758 | 0.790 | 0.763 | 0.648 | 0.740 0.221 0.221 |
Ours ( ) ) |
0.696 | 0.725 | 0.729 | 0.699 | 0.638 | 0.697 0.204 0.204 |
0.744 | 0.756 | 0.796 | 0.752 | 0.654 | 0.739 0.213 0.213 |
Ours (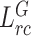 + +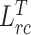 ) ) |
0.723 | 0.728 | 0.718 | 0.720 | 0.625 |
0.703 0.193 0.193
|
0.756 | 0.760 | 0.790 | 0.771 | 0.631 |
0.742 0.203 0.203
|
Fig. 3.

Visual comparison of COVID-19 infection segmentation by different methods on 3D COVID-19 segmentation benchmark dataset from axial view.
E. 3D Comparison Experiments on COVID-19 Segmentation Benchmark Dataset
To demonstrate the effectiveness of our method, we conduct extensive comparison experiments with other state-of-the-art methods. To ensure a fair comparison, all methods are experimented with the same network backbone and experimental settings. Segmentation models trained from scratch with only COVID-19 cases serve as our baseline results. Besides, as a simple and intuitive approach, pre-training segmentation models on non-COVID cases and fine-tuning on COVID-19 cases are utilized as comparison results for learning from both COVID-19 and non-COVID cases. The quantitative experimental results are shown in Table IV. From the results, we can observe that transferring pre-trained models to COVID-19 infection segmentation tasks can generally improve the performance of training from scratch with only COVID-19 cases on most experiments, and using multi-lesion is superior to single-lesion when more general representations can be utilized to help COVID-19 infection tasks, with 2.3% and 1.7% improvements in DSC and NSD, respectively. However, these transfer learning methods show instability under different data distribution in five-fold cross validation experiments. The rationale is that the transfer ability largely depends on the domain difference between datasets. When there exists a large domain distance between non-COVID and limited COVID-19 training cases, transfer learning may somehow mislead the learning procedure.
TABLE IV. Quantitative Results of 5-Fold Cross Validation of 3D Comparison Experiments With State-of-the-Art Methods.
| Method | DSC | NSD | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fold 0 | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Avg | Fold 0 | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Avg | |
| 3D nnUNet | 0.681 | 0.713 | 0.662 | 0.681 | 0.627 | 0.673 0.223 0.223 |
0.709 | 0.718 | 0.717 | 0.708 | 0.649 | 0.700 0.224 0.224 |
| Pre-train on MSD | 0.679 | 0.706 | 0.724 | 0.708 | 0.623 | 0.688 0.201 0.201 |
0.706 | 0.708 | 0.785 | 0.724 | 0.642 | 0.713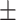 0.225 0.225 |
| Pre-train on NSCLC | 0.696 | 0.716 | 0.673 | 0.690 | 0.579 | 0.671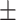 0.228 0.228 |
0.714 | 0.720 | 0.734 | 0.707 | 0.590 | 0.693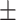 0.248 0.248 |
Pre-train on Multi-lesion
|
0.702 | 0.736 | 0.703 | 0.725 | 0.612 | 0.696 0.213 0.213 |
0.715 | 0.730 | 0.755 | 0.754 | 0.628 | 0.717 0.227 0.227 |
Multi-encoder
|
0.712 | 0.732 | 0.721 | 0.742 | 0.608 | 0.703 0.201 0.201 |
0.735 | 0.740 | 0.785 | 0.772 | 0.629 | 0.732 0.218 0.218 |
| Ours-3D | 0.723 | 0.728 | 0.718 | 0.720 | 0.625 | 0.703 0.193 0.193 |
0.756 | 0.760 | 0.790 | 0.771 | 0.631 | 0.742 0.203 0.203 |
5Note:  denotes the results from [18] where additional non-COVID datasets are used.
denotes the results from [18] where additional non-COVID datasets are used.
The best results are shown in red font and the second-best results in blue font.
In [18], the authors propose a multi-encoder architecture to freeze the non-COVID pre-trained encoder as an additional feature extractor for the training of COVID-19 cases. Features from the frozen adapted-encoder and reinitialized self-encoder are concatenated for the subsequent decoder. However, their workflow is still based on transfer learning, that training a network first on non-COVID cases and then on COVID-19 cases with foregoing pre-trained parameters. The main limitation is that the learning procedures of two tasks are separate. Therefore, the shared knowledge of non-COVID and COVID-19 cases cannot be fully exploited. It is observed that our method takes advantage of collaborative learning between two encoders and interactively improves the overall learning procedure. As a consequence, our method achieves higher segmentation performance with an averaged DSC of 70.3% and averaged NSD of 74.2%. Comparing with training from scratch, exploiting shared knowledge from non-COVID lesions can achieve further improvements with up to 3.0% in DSC and 4.2% in NSD. Paired T-test shows that the improvements are statistically significant at  , validating the effectiveness of our proposed method.
, validating the effectiveness of our proposed method.
F. 2D Comparison Experiments on COVID-SemiSeg Dataset
In this subsection, we compare our method with state-of-the-art methods for 2D medical image segmentation, including U-Net [37], U-Net++ [50], Dense-UNet [49], Attention-UNet [47], Gated-UNet [48], Inf-Net and Semi-Inf-Net [30]. Quantitative results are shown in Table V. As can be observed, our proposed method outperforms all comparing methods on all evaluation metrics by a large margin, validating the effectiveness of our framework. Paired T-test shows that the improvements are statistically significant at  . Besides, we visualize some segmentation results of our method in Fig. 4. These results indicate that our segmentation results are closer to the ground truth with less mis-segmented areas and outperform other methods significantly. For semi-supervised setting, we additionally integrate unlabeled COVID-19 cases of COVID-SemiSeg Dataset into the relation-driven training in our framework. These unlabeled cases can be utilized for the regularization of feature relation to achieve more consistent and robust learning and further improve the segmentation performance slightly.
. Besides, we visualize some segmentation results of our method in Fig. 4. These results indicate that our segmentation results are closer to the ground truth with less mis-segmented areas and outperform other methods significantly. For semi-supervised setting, we additionally integrate unlabeled COVID-19 cases of COVID-SemiSeg Dataset into the relation-driven training in our framework. These unlabeled cases can be utilized for the regularization of feature relation to achieve more consistent and robust learning and further improve the segmentation performance slightly.
TABLE V. Quantitative Results of 2D Comparison Experiments With State-of-the-Art Methods for Fully Supervised Segmentation and Semi-Supervised Segmentation.
| Methods | Dice | Sensitivity | Specificity | MAE |
|---|---|---|---|---|
| U-Net [37] | 0.439 | 0.534 | 0.858 | 0.186 |
| Attention-UNet [47] | 0.583 | 0.637 | 0.921 | 0.112 |
| Gated-UNet [48] | 0.623 | 0.658 | 0.926 | 0.102 |
| Dense-UNet [49] | 0.515 | 0.594 | 0.840 | 0.184 |
| U-Net++ [50] | 0.581 | 0.672 | 0.902 | 0.120 |
| Inf-Net [30] | 0.682 | 0.692 | 0.943 | 0.082 |
| Semi-Inf-Net [30] | 0.739 | 0.725 | 0.960 | 0.064 |
| 2D nnUNet [45] | 0.747 | 0.829 | 0.953 | 0.063 |
| Ours-2D | 0.765 | 0.839 | 0.955 | 0.056 |
| Ours-2D-UN | 0.767 | 0.839 | 0.957 | 0.055 |
Fig. 4.
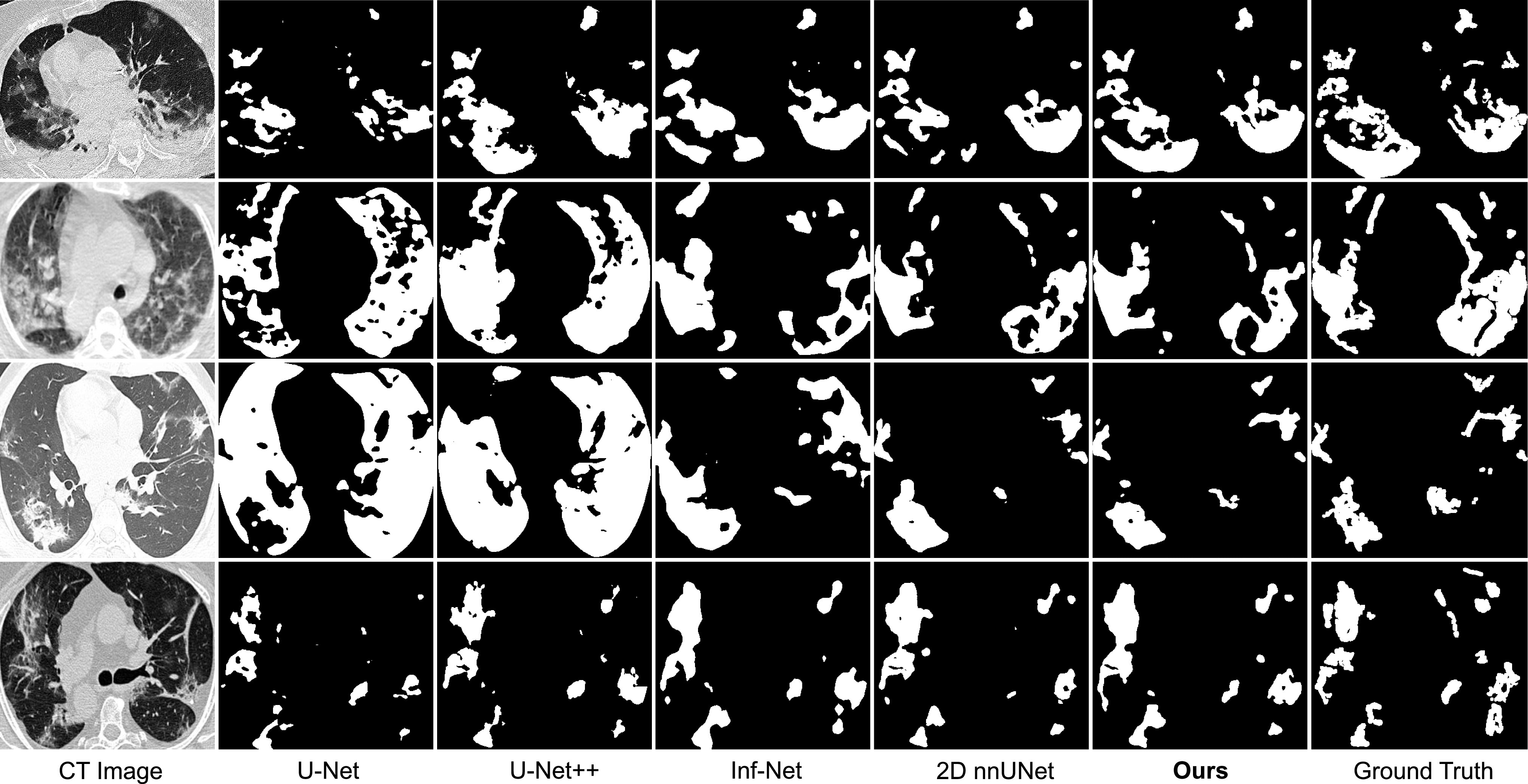
Visual comparison of COVID-19 infection segmentation by different methods on 2D COVID-SemiSeg dataset. As can be observed, our method can generate segmentation results with more accurate boundaries and less segmentation mistakes in small infection areas, which is closer to the ground truth.
G. Comparison on Different Datasets for Shared Knowledge Learning
To better demonstrate the effectiveness of our proposed feature relation-driven learning, we make extensive experiments on several lesion segmentation datasets of medical images with different relation to COVID-19 infections. In addition to the lung tumor and pleural effusion datasets introduced before, we use LiTS dataset [9] with liver tumor annotations in abdominal CT volumes as non-COVID lesions in our method for comparison. Besides, to make comparison between intra-disease and inter-disease relations, we use another multi-national CT dataset with labeled ground glass opacities [51] as an out-of-domain dataset for the learning of general branch, which is more relevant with similar appearance to the target dataset in our framework.
In our experiments, we follow the settings of our 2D experiments with the same network backbone and implementation details. To make quantitative comparisons, we select out the same amount of 60 cases from these different datasets for the general branch, and 10 cases from the COVID-19 segmentation dataset for target branch. The experimental results are shown in Table VI. It can be observed from the table that using intra-disease dataset that are more related to the target dataset can achieve better performance compared with other datasets under the same condition, which proves that more similar appearance can lead to more significant improvement by exploiting shared knowledge. Specifically, we observe that using non-lung lesions can also obtain comparable results compared with experiments using non-COVID lung lesions like lung tumor and pleural effusion.
TABLE VI. Quantitative Comparison Using Different Datasets as non-COVID Cases for Shared Knowledge Learning.
| Dataset / description | Dice | Sensitivity | Specificity | MAE |
|---|---|---|---|---|
| None / COVID only | 0.693 | 0.774 | 0.941 | 0.080 |
| LiTS / liver tumor | 0.711 | 0.782 | 0.942 | 0.074 |
| MSD Lung / lung nodules | 0.712 | 0.764 | 0.945 | 0.073 |
| NSCLC / pleural effusion | 0.702 | 0.781 | 0.941 | 0.072 |
| COVID-19-20 / groundglass opacities | 0.714 | 0.792 | 0.942 | 0.070 |
All the experiments are performed with the same amount of 60 non-COVID cases and 10 COVID-19 cases under the same settings
H. Visual Analysis of Our Method
To visualize learning procedure of our method, we show some examples of general and target feature relation matrices at different epochs during the network training procedure in Fig. 5 and Fig. 6. The absolute differences of these two matrices are shown in the right column in red to clearly visualize the alignment of matrices.It can be observed in Fig. 5 that as the training goes on, the general encoder gradually produces relation matrices with higher response at the same channel. Meanwhile, the absolute differences of feature relation matrices of non-COVID and COVID-19 cases extracted from general encoder are gradually decreased, indicating that the general encoder learns more general and robust representations of lung lesions.Besides, as observed in Fig. 6, the absolute differences of general and target feature relation matrices of COVID-19 cases are gradually increased and tend to be stable as the training goes on, indicating that the target encoder is gradually enforced to focus on task-specific features and learn more discriminative representations compared with general encoder.
Fig. 5.
Visualization of the general feature relation matrices of non-COVID cases (left column) and COVID-19 cases (middle column) and their absolute difference (right column) during the training procedure.
Fig. 6.
Visualization of the general feature relation matrix (left column) and target feature relation matrix (middle column) of COVID-19 cases and their absolute difference (right column) during the training procedure.
V. Discussion
With the outbreak of COVID-19 all over the world, designing effective automated tools for fighting against COVID-19 is highly demanded to improve the efficiency of clinical approaches and reduce the tedious workload of clinicians and radiologists. However, accurate segmentation of COVID-19 lung infections is a challenging task due to the large appearance variance of COVID-19 lesions of patients in different severity level, and existing data-driven segmentation methods mainly rely on large amount of well annotated data. In order to mitigate the insufficiency of labeled COVID-19 CT scans, it is essential and meaningful to develop annotation-efficient segmentation methods for the COVID-19 lung infection segmentation task.
Considering that there are several public non-COVID lung lesion segmentation datasets due to other clinical practice, these datasets may serve as potential profit for generalizing useful information to assist in the related COVID-19 infection segmentation task. Some previous studies also highlight the usage of non-COVID lung lesions [18], [42]. However, these existing approaches merely focus on investigating the transferability in COVID-19 infection segmentation. Although their results reveal benefits of pre-training on non-COVID datasets, the improvement is limited when shared knowledge between COVID-19 and non-COVID lung lesions cannot be fully utilized. Our experiment reveal that the proposed collaborative learning scheme can effectively exploit shared semantic information by regularizing the consistency between extracted features and promote the training procedure in the absence of sufficient high-quality COVID-19 data. In addition, our scheme can be extended to utilize unlabeled COVID-19 data for feature relation regularization. Experimental results show that even without annotations, our method can use unlabeled scans to explore feature relation and achieve more consistent and robust learning. Fig. 7 presents an example of challenging cases for COVID-19 lung infection segmentation. Although our method can achieve significant improvement by exploiting knowledge from non-COVID lesions, the limitation still exists. We observe that comparing with ground truth, there are still some mis-segmented areas when encountering challenging cases with multiple irregular infections. As a near future work, we intend to explore how to achieve more robust and reliable knowledge transfer. In addition, we also plan to extend our method to other medical image segmentation tasks to explore the usage of out-of-domain datasets for annotation-efficient deep learning, thus enhancing the applicability of these methods in real-world applications.
Fig. 7.
Example of challenging cases for COVID-19 lung infection segmentation with limited labeled data.
VI. Conclusion
In this paper, we propose a novel relation-driven collaborative learning model to exploit shared knowledge from non-COVID lesions for annotation-efficient COVID-19 CT lung infection segmentation. Specifically, the model consists of two encoders with the same architecture and a shared decoder. The general encoder is adopted to capture general lung lesion features based on multiple non-COVID lesions and the target encoder is adopted to focus on task-specific feature of COVID-19 infection. To exploit shared knowledge from non-COVID lesions, we develop a collaborative learning scheme to regularize the relation between extracted features of given input for the training. We present a set of experiments on 2D slices and 3D volumes based on three COVID-19 datasets and two non-COVID datasets. Experimental results reveal clear benefits of utilizing non-COVID lesions in the absence of sufficient COVID-19 annotations to train a robust segmentation model. Moreover, we provide a semi-supervised learning solution to utilize the unlabeled COVID-19 cases for feature relation regularization and achieved performance improvements. Among all comparison experiments, our proposed method outperforms state-of-the-art methods and illustrates strong potential for real-world applications in the global fight against COVID-19.
Funding Statement
This work was supported in part by the National Key Research and Development Program of China under Grant 2016YFF0201002, in part by the University Synergy Innovation Program of Anhui Province under Grant GXXT-2019-044, and in part by the National Natural Science Foundation of China under Grant 61301005.
Footnotes
[Online]. Avilable: https://coronavirus.jhu.edu/map.html.
[Online]. Available: https://zenodo.org/record/3757476#.X4ABeYvivid.
[Online]. Available: http://medicalsegmentation.com/covid19/.
[Online]. Available: http://medicaldecathlon.com/.
[Online]. Available: https://wiki.cancerimagingarchive.net/display/Public/NSCLC-Radiomics.
Contributor Information
Yichi Zhang, Email: coda1998@buaa.edu.cn.
Qingcheng Liao, Email: liaoqingcheng@buaa.edu.cn.
Lin Yuan, Email: a11111good@163.com.
He Zhu, Email: roy_zh@buaa.edu.cn.
Jiezhen Xing, Email: xingjiezhen@buaa.edu.cn.
Jicong Zhang, Email: jicongzhang@buaa.edu.cn.
References
- [1].Mazzone P. J. et al. , “Management of lung nodules and lung cancer screening during the COVID-19 pandemic: CHEST expert panel report,” Chest, vol. 158, no. 1, pp. 406–415, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Oudkerk M. et al. , “Diagnosis, prevention, and treatment of thromboembolic complications in COVID-19: Report of the national institute for public health of the Netherlands,” Radiology, vol. 297, no. 1, pp. E216–E222, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li Y. and Xia L., “Coronavirus disease 2019 (COVID-19): Role of chest CT in diagnosis and management,” Amer. J. Roentgenol., vol. 214, no. 6, pp. 1280–1286, 2020. [DOI] [PubMed] [Google Scholar]
- [4].Chung M. et al. , “CT imaging features of 2019. novel coronavirus (2019-NCOV),” Radiology, vol. 295, no. 1, pp. 202–207, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fang Y. et al. , “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. E115–E117, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li K. et al. , “CT image visual quantitative evaluation and clinical classification of coronavirus disease (COVID-19),” Eur. Radiol., vol. 30, no. 8, pp. 4407–4416, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shi F. et al. , “Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19,” IEEE Rev. Biomed. Eng., vol. 14, pp. 4–15, 2020. [DOI] [PubMed] [Google Scholar]
- [8].Joskowicz L., Cohen D., Caplan N., and Sosna J., “Inter-observer variability of manual contour delineation of structures in CT,” Eur. Radiol., vol. 29, no. 3, pp. 1391–1399, 2019. [DOI] [PubMed] [Google Scholar]
- [9].Bilic P. et al. , “The liver tumor segmentation benchmark (LITS),” 2019, arXiv:1901.04056. [DOI] [PMC free article] [PubMed]
- [10].Heller N. et al. , “The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge,” Med. Image Anal., vol. 67, 2019, Art. no. 101821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bernard O. et al. , “Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved?,” IEEE Trans. Med. Imag., vol. 37, no. 11, pp. 2514–2525, Nov. 2018. [DOI] [PubMed] [Google Scholar]
- [12].Ma J. et al. , “Abdomenct-1k: Is abdominal organ segmentation a solved problem?,” IEEE Trans. Pattern Anal. Mach. Intell., 2021, to be published, doi: 10.1109/TPAMI.2021.3100536. [DOI] [PubMed]
- [13].Huang L. et al. , “Serial quantitative chest CT assessment of COVID-19: Deep-learning approach,” Radiol., Cardiothoracic Imag., vol. 2, no. 2, 2020, Art. no. e200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun L. et al. , “Adaptive feature selection guided deep forest for COVID-19 classification with chest CT,” IEEE J. Biomed. Health Informat., vol. 24, no. 10, pp. 2798–2805, Oct. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang K. et al. , “Clinically applicable ai system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography,” Cell, vol. 181, no. 6, pp. 1423–1433, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Z., Liu Q., and Dou Q., “Contrastive cross-site learning with redesigned net for COVID-19 CT classification,” IEEE J. Biomed. Health Informat., vol. 24, no. 10, pp. 2806–2813, Oct. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ouyang X. et al. , “Dual-sampling attention network for diagnosis of COVID-19 from community acquired pneumonia,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2595–2605, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [18].Wang Y. et al. , “Does non-COVID19 lung lesion help? investigating transferability in COVID-19 CT image segmentation,” Comput. Methods and Programs in Biomedicine, vol. 202, p. 106004, 2021, arXiv:2006.13877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cheplygina V., Bruijne M. de, and Pluim J. P., “Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis,” Med. Image Anal., vol. 54, pp. 280–296, 2019. [DOI] [PubMed] [Google Scholar]
- [20].Tajbakhsh N., Jeyaseelan L., Li Q., Chiang J. N., Wu Z., and Ding X., “Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation,” Med. Image Anal., vol. 63, 2020, Art. no. 101693. [DOI] [PubMed] [Google Scholar]
- [21].Engelen J. E. Van and Hoos H. H., “A survey on semi-supervised learning,” Mach. Learn., vol. 109, no. 2, pp. 373–440, 2020. [Google Scholar]
- [22].Ji Z., Shen Y., Ma C., and Gao M., “Scribble-based hierarchical weakly supervised learning for brain tumor segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervention, 2019, pp. 175–183. [Google Scholar]
- [23].Huang R., Zheng Y., Hu Z., Zhang S., and Li H., “Multi-organ segmentation via co-training weight-averaged models from few-organ datasets,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervention, 2020, pp. 146–155. [Google Scholar]
- [24].Zhang J., Xie Y., Xia Y., and Shen C., “DodNet: Learning to segment multi-organ and tumors from multiple partially labeled datasets,” in Proc. IEEE/CVF Conf. Comput. Vision and Pattern Recognition, 2021, pp. 1195–1204. [Google Scholar]
- [25].Shan F. et al. , “Lung infection quantification of COVID-19 in CT images with deep learning,” 2020, arXiv:2003.04655.
- [26].Amyar A., Modzelewski R., Li H., and Ruan S., “Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation,” Comput. Biol. Med., vol. 126, 2020, Art. no. 104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xie W., Jacobs C., Charbonnier J.-P., and Ginneken B. Van, “Relational modeling for robust and efficient pulmonary lobe segmentation in CT scans,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2664–2675, Aug. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhou T., Canu S., and Ruan S., “An automatic COVID-19 CT segmentation network using spatial and channel attention mechanism,” 2020, arXiv:2004.06673. [DOI] [PMC free article] [PubMed]
- [29].Zheng C. et al. , “Deep learning-based detection for COVID-19 from chest CT using weak label,” MedRxiv, 2020. [Google Scholar]
- [30].Fan D.-P. et al. , “Inf-Net: Automatic COVID-19 lung infection segmentation from CT images,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2626–2637, Aug. 2020. [DOI] [PubMed] [Google Scholar]
- [31].Wang G. et al. , “A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images,” IEEE Trans. Med. Imag., vol. 39, no. 8, pp. 2653–2663, Jun. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yao Q., Xiao L., Liu P., and Zhou S. K., “Label-free segmentation of COVID-19 lesions in lung CT,” IEEE Trans. Medical Imaging, 2021, arXiv:2009.06456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ma J. et al. , “Active contour regularized semi-supervised learning for COVID-19 CT infection segmentation with limited annotations,” Phys. Med. Biol., vol. 65, no. 22, 2020, Art. no. 225034. [DOI] [PubMed] [Google Scholar]
- [34].Chouhan V. et al. , “A novel transfer learning based approach for pneumonia detection in chest X-ray images,” Appl. Sci., vol. 10, no. 2, 2020, Art. no. 559. [Google Scholar]
- [35].Majeed T., Rashid R., Ali D., and Asaad A., “COVID-19 detection using CNN transfer learning from X-ray images,” Medrxiv, 2020. [Google Scholar]
- [36].Misra S., Jeon S., Lee S., Managuli R., Jang I.-S., and Kim C., “Multi-channel transfer learning of chest X-ray images for screening of COVID-19,” Electron., vol. 9, no. 9, 2020, Art. no. 1388. [Google Scholar]
- [37].Ronneberger O., Fischer P., and Brox T., “U-Net: Convolutional networks for biomedical image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervention, 2015, pp. 234–241. [Google Scholar]
- [38].Çiçek Ö., Abdulkadir A., Lienkamp S. S., Brox T., and Ronneberger O., “3D U-Net: Learning dense volumetric segmentation from sparse annotation,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervention, 2016, pp. 424–432. [Google Scholar]
- [39].Liu Q., Yu L., Luo L., Dou Q., and Heng P. A., “Semi-supervised medical image classification with relation-driven self-ensembling model,” IEEE Trans. Med. Imag., vol. 39, no. 11, pp. 3429–3440, Nov. 2020. [DOI] [PubMed] [Google Scholar]
- [40].Gatys L., Ecker A., and Bethge M., “A neural algorithm of artistic style,” J. Vis., vol. 16, no. 12, pp. 326–326, 2016. [Google Scholar]
- [41].Dou Q., Chen H., Jin Y., Yu L., Qin J., and Heng P.-A., “3D deeply supervised network for automatic liver segmentation from CT volumes,” in Proc. Int. Conf. Med. Image Comput. Comput.-Assist. Intervention, 2016, pp. 149–157. [Google Scholar]
- [42].Ma J. et al. , “Towards data-efficient learning: A benchmark for COVID-19 CT lung and infection segmentation,” Med. Phys., vol. 48, no. 3, pp. 1197–1210, 2020. [DOI] [PubMed] [Google Scholar]
- [43].Simpson A. L. et al. , “A large annotated medical image dataset for the development and evaluation of segmentation algorithms,” 2019, arXiv:1902.09063.
- [44].Paszke A. et al. , “PyTorch: An imperative style, high-performance deep learning library,” in Proc. Adv. Neural Inf. Process. Syst., 2019, pp. 8026–8037. [Google Scholar]
- [45].Isensee F., Jaeger P. F., Kohl S. A., Petersen J., and Maier-Hein K. H., “nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation,” Nature Meth., vol. 18, no. 2, pp. 203–211, 2020. [DOI] [PubMed] [Google Scholar]
- [46].Laine S. and Aila T., “Temporal ensembling for semi-supervised learning,” 2016, arXiv:1610.02242.
- [47].Oktay O. et al. , “Attention U-Net: Learning where to look for the pancreas,” 2018, arXiv:1804.03999.
- [48].Schlemper J. et al. , “Attention gated networks: Learning to leverage salient regions in medical images,” Med. Image Anal., vol. 53, pp. 197–207, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li X., Chen H., Qi X., Dou Q., Fu C.-W., and Heng P.-A., “H-denseUNet: Hybrid densely connected unet for liver and tumor segmentation from CT volumes,” IEEE Trans. Med. Imag., vol. 37, no. 12, pp. 2663–2674, Dec. 2018. [DOI] [PubMed] [Google Scholar]
- [50].Zhou Z., Siddiquee M. M. R., Tajbakhsh N., and Liang J., “UNet++: Redesigning skip connections to exploit multiscale features in image segmentation,” IEEE Trans. Med. Imag., vol. 39, no. 6, pp. 1856–1867, Jun. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Roth H. et al. , “Rapid artificial intelligence solutions in a pandemic-the COVID-19-20 lung CT lesion segmentation challenge,” Res. Sq., 2021, doi: 10.21203/rs.3.rs-571332/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]