Academia.edu no longer supports Internet Explorer.
To browse Academia.edu and the wider internet faster and more securely, please take a few seconds to upgrade your browser.
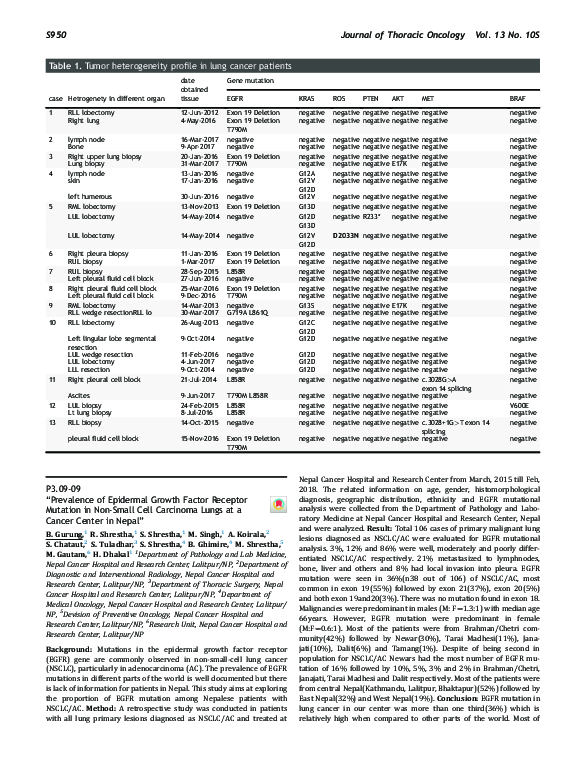
P3.09-09 “Prevalence of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Carcinoma Lungs at a Cancer Center in Nepal”
P3.09-09 “Prevalence of Epidermal Growth Factor Receptor Mutation in Non-Small Cell Carcinoma Lungs at a Cancer Center in Nepal”
2018, Journal of Thoracic Oncology
Related Papers
Annals of Oncology
Epidermal growth factor receptor activating mutations in Spanish gefitinib-treated non-small-cell lung cancer patients2005 •
Indian Journal of Medical and Paediatric Oncology
Epidermal growth factor receptor mutation in adenocarcinoma lung in a North Indian population: Prevalence and relation with different clinical variables2016 •
Introduction: Lung cancer is one of the most common causes of cancer deaths worldwide. Adenocarcinoma is taking over squamous cell lung cancer as the predominant histological subtype. Several cytotoxic drugs are available for the treatment of lung cancer, but side effects limit their use. Recently, targeted therapies for cancers have come into clinical practice. Aims and Objectives: To determine the prevalence of epidermal growth factor receptor (EGFR) mutation in adenocarcinoma lung in a North Indian population and its relation with different clinical variables. Materials and Methods: A total of 57 patients who met inclusion criteria were recruited into the study. Relevant history, clinical examination and investigations were done. EGFR mutation was done in all patients. Results: A total of twenty patients tested positive for EGFR mutation. EGFR was more frequently detected in female patients (53.8%), while as only 19.4% of the male patients expressed EGFR mutation, which was stati...
Lung Cancer (Auckland, N.Z.)
{"__content__"=>"Uncommon mutations in cytological specimens of 1,874 newly diagnosed Indonesian lung cancer patients.", "i"=>{"__content__"=>"EGFR"}}2018 •
We aimed to evaluate the distribution of individual epidermal growth factor receptor () mutation subtypes found in routine cytological specimens. A retrospective audit was performed on testing results of 1,874 consecutive cytological samples of newly diagnosed or treatment-naïve Indonesian lung cancer patients (years 2015-2016). Testing was performed by ISO15189 accredited central laboratory. Overall test failure rate was 5.1%, with the highest failure (7.1%) observed in pleural effusion and lowest (1.6%) in needle aspiration samples. mutation frequency was 44.4%. Tyrosine kinase inhibitor (TKI)-sensitive common mutations (ins/dels exon 19, L858R) and uncommon mutations (G719X, T790M, L861Q) contributed 57.1% and 29%, respectively. Approximately 13.9% of mutation-positive patients carried a mixture of common and uncommon mutations. Women had higher mutation rate (52.9%) vs men (39.1%; <0.05). In contrast, uncommon mutations conferring either TKI responsive (G719X, L861Q) or TKI r...
2010 •
Journal of Thoracic Disease
Detection of epidermal growth factor receptor mutations in circulating tumor DNA: reviewing BENEFIT clinical trial2018 •
Molecular and Clinical Oncology
Real‑world evaluation of molecular testing and treatment patterns for EGFR mutations in non‑small cell lung cancer in Latin America2021 •
International Journal of Research and Development in Pharmacy and Life Sciences
Epidermal Growth Factor Receptor Mutations detection by Mutant Specific Immunohistochemistry in North Indian Lung Cancer population2017 •
http://dx.doi.org/10.21276/IJRDPL.227 8-0238.2017.6(6).2796-2799 ABSTRACT: Background: Lung cancer is one of the leading causes of cancer-related deaths due to limited treatment options available for advanced-stage disease. The epidermal growth factor receptor is a trans-membrane glycoprotein having an extracellular epidermal growth factor binding domain and an intracellular tyrosine kinase domain which regulates signaling pathways to control cellular proliferation. Material and Methods: Detection of EGFR mutation status of 80 subjects of non-small cell lung cancer of adenocarcinoma subtype had been done by IHC using EGFR mutation specific antibodies. Results: 20 cases were positive for Del E746-A750 mutation, 6 for L858R mutation. IHC analysis shows that 30 cases scored 0, 12 cases scored 2+ and 8 cases scored 3+ in Del E746-A750 mutation while 24 cases scored 0, 1 case scored 2+ and 5 cases scored 3+ in L858R mutation. The mean age of mutation positive is 56±9.50 and negative is 5...
Indian Journal of Cancer
Epidermal growth factor receptor mutation subtypes and geographical distribution among Indian non-small cell lung cancer patients2013 •
Journal of Cancer Research and Therapeutics
Frequency of epidermal growth factor receptor mutations in Jordanian lung adenocarcinoma patients at diagnosis2016 •
2024 •
In the past, most iconographical studies on slavery and similar phenomena focused on specific regions, cultures and periods. The aim of this conference is to look at a broad range of dependent and marginalized social groups and 'others' and to compare the results of iconographical studies on different pre-modern societies (prior to 1800 CE) around the globe. Therefore, we invited scholars from a wide variety of disciplines (Near Eastern Archaeology, Egyptology, Classical Archaeology, European Art History, Asian Art History, Anthropology of the Americas) in order to gain new insights by using diachronic and cross-cultural comparisons.
RELATED PAPERS
Ciência política e governança global 2 (Atena Editora)
Ciência política e governança global 2 (Atena Editora)2024 •
www.eduardorojotorrecilla.es
Cataluña. Inmigración. Los deseos de algunos y la realidad jurídica2024 •
Pneumonologia i Alergologia Polska
Czynniki określające sukces w porzucaniu palenia u osób uczestniczących w środowiskowych badaniach spirometrycznychRevista da Faculdade de Direito UFPR
APROXIMAÇÃO CRÍTICA ENTRE AS JURISDIÇÕES DE CIVIL LAW E DE COMMON LAW E A NECESSIDADE DE RESPEITO AOS PRECEDENTES NO BRASIL2010 •
Chronique internationale de l'IRES
Belgique. Hausse des salaires nominaux, aides publiques ponctuelles et perte de pouvoir d’achat2023 •
Revue médicale de Liège
Recommandations Europeennes concernant le diagnostic de l'embolie pulmonaire2015 •
2014 •
Medicina Fluminensis
Primjena imunoterapije kod onkoloških bolesnika s bolešću COVID-192022 •
Archives of Toxicology
Multiparametric assessment of mitochondrial respiratory inhibition in HepG2 and RPTEC/TERT1 cells using a panel of mitochondrial targeting agrochemicals2020 •
Advances in Genetics Volume 80
Precision Editing of Large Animal Genomes2012 •
2019 •
RELATED TOPICS
- Find new research papers in:
- Physics
- Chemistry
- Biology
- Health Sciences
- Ecology
- Earth Sciences
- Cognitive Science
- Mathematics
- Computer Science
 Banita Gurung
Banita Gurung