Abstract
It is apparent that various functional units within the cellular machinery are derived from RNAs. The evolution of sequencing techniques has resulted in significant insights into approaches for transcriptome studies. Organisms utilize RNA to govern cellular systems, and a heterogeneous class of RNAs is involved in regulatory functions. In particular, regulatory RNAs are increasingly recognized to participate in intricately functioning machinery across almost all levels of biological systems. These systems include those mediating chromatin arrangement, transcription, suborganelle stabilization, and posttranscriptional modifications. Any class of RNA exhibiting regulatory activity can be termed a class of regulatory RNA and is typically represented by noncoding RNAs, which constitute a substantial portion of the genome. These RNAs function based on the principle of structural changes through cis and/or trans regulation to facilitate mutual RNAâRNA, RNAâDNA, and RNAâprotein interactions. It has not been clearly elucidated whether regulatory RNAs identified through deep sequencing actually function in the anticipated mechanisms. This review addresses the dominant properties of regulatory RNAs at various layers of the cellular machinery and covers regulatory activities, structural dynamics, modifications, associated molecules, and further challenges related to therapeutics and deep learning.
Similar content being viewed by others
Introduction
Regulatory RNAs exhibit highly dynamic properties due to their multidimensional structures and their ability to interact with RNA, DNA, and proteins. These RNAs are involved in regulated events, such as transcription, translation, molecular localization and stability, and enzymatic degradation cascades, on many levels1,2,3,4. A broad spectrum of RNAs that have regulatory functions are collectively recognized as regulatory RNAs. Historically, many tiers of noncoding RNAs (ncRNAs) have been identified that have diverse characteristics. These ncRNAs account for approximately 98% of the total transcriptome, and while some ncRNAs exhibit protein-coding ability5, the majority of ncRNAs are expected to function as regulatory RNAs6,7. Regulatory RNAs can be classified into different groups based on their size, structure, localization, and function, as indicated by the discovery of long noncoding RNAs (lncRNAs), enhancer RNAs (eRNAs), microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), small Cajal body-specific RNAs (scaRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), and circular RNAs (circRNAs)7,8. The enormous functional involvement of regulatory RNAsâdespite their low expression level and low sequence conservationâis undeniable, and they apparently play important roles in cellular phenomena through these functions9. Research on RNAs and their biogenesis has described both shared and distinctive features among the numerous types of regulatory RNAs. Numerous regulatory RNAs undergo mRNA-like RNA processing steps, such as 5â² m7G cap addition, polyadenylation, and splicing, which are observed even in long RNAs. The pattern of overlapping transcripts in the genome indicates the inclusion or exclusion of exons and introns in the sequences of regulatory RNAs, which influences the open reading frame10. Additionally, the directionality of transcription is can be either unidirectional or bidirectional, possibly implying base pairing-dependent functions11. LncRNAs, with more than 200 nucleotides, are the most common type of RNA undergoing mRNA-like processes, and the lncRNA class contains numerous kinds of regulatory RNAs due to the arbitrary definition of this category considering only length. Numerous lncRNAs are involved in multiple mechanisms in multiple cellular components. They interact with other RNA molecules or proteins, thereby modulating cellular signaling and epigenetic regulation12,13,14,15. Active enhancer sequence-derived eRNAs are considerably lengthy, i.e., more than 1 kilobase, and are responsible for enhancer-associated contributions16,17. Given this knowledge, miRNAs are also commonly referenced regulatory RNA class. The biogenesis of regulatory miRNAs involves several sequential steps. A hairpin-structured primary miRNA (pri-miRNA) is cleaved by the RNase III Drosha to a length of 55â70 nucleotides in the nucleus; the resulting pre-miRNA is released into the cytoplasm and is then cleaved into a miRNA duplex by the RNase III Dicer. Mature miRNAs bind to complementary mRNAs, repressing translation through an interaction in the 3â² untranslated region (UTR) of the target mRNA. The RNA-induced silencing complex (RISC) and argonaute cooperate to actively regulate target interference18,19,20. Similar to miRNAs, piRNAs utilize argonaute but are distinguished by their sequences, mainly derived from transposons, and are comparatively more involved in epigenetic regulatory processes21. Regulatory RNAs found in nucleoli, termed snoRNAs, have a unique set of sequences that allow their subclassification into C/D box snoRNAs (SNORDs), H/ACA box snoRNAs (SNORAs), and far more specific localizing motifs define scaRNAs found only in Cajal bodies. These motifs lead to 2â²-O-methylation and pseudouridylation modifications in other classes of RNA, conferring regulatory identity22,23. Another unique class, circRNAs, possess a closed circular structure formed by back-splicing of pre-mRNAs, unlike the linear form of other regulatory RNAs. Their splicing determines the location of their function; exonic circRNAs are transported to the cytoplasm, whereas intronic and intro-exon circRNAs remain in the nucleus24,25.
Currently, research is at an intersection defined by exponentially increasing numbers of datasets generated through high-throughput sequencing techniques26. This surge is essential for genomic research, especially in defining the pool of regulatory RNAs. For instance, RNA sequencing and chromatin immunoprecipitation sequencing aided in enabling eRNA discovery and investigation27,28. Multiple kinds of sequencing methods have been adopted to examine regulatory RNAs in detail with procedural commonalities, from RNA preparation to analysis of aligned sequences (Table 1). Numerous efforts have been made to determine the regulatory functions of RNAs29,30. The interaction of regulatory RNAs with accompanying molecules has been a key factor in designing sequencing methods. RNA-associated chromosomal conformation studies can be conducted with techniques such as Hi-C coupled with chromatin isolation by RNA purification, chromatin-associated RNA sequencing, and RNA and DNA interacting complexes ligated and sequenced31,32,33,34. To identify proteins that specifically bind to RNA, methods such as UV crosslinking with immunoprecipitation2,35,36, photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation2,37,38, and individual-nucleotide resolution UV crosslinking and immunoprecipitation can be adapted2,39,40. As alterations in RNA structure directly affect RNA function, sequencing methods such as RNA in situ conformation sequencing (RIC-seq)41,42, mapping RNA interactome in vivo43, and dimethyl sulfate mutational profiling with sequencing44,45,46 can be utilized to investigate such RNA structure-dependent aspects. Additionally, the RNA:DNA hybridization-mediated structure and its attributes can be confirmed by DNAâRNA immunoprecipitation coupled with high-throughput sequencing (DRIP-seq)47,48,49, DNA:RNA immunoprecipitation followed by cDNA conversion coupled with high-throughput sequencing (DRIPc-seq)47,50 and bisulfite-based DRIP-seq51. More specifically, single-molecule structure sequencing can be used to investigate tertiary interactions, the dynamics of riboswitch ligand binding, and mRNA structural features1. With respect to the subcellular localization of regulatory RNAs, spatial transcripts perform distinct functions across tissues according to the spatial context. Advanced single-cell sequencing techniques allow researchers to access highly resolved spatial transcriptomes at intercellular resolution without external perturbation52,53,54. Databases focusing on regulatory RNAs, such as lncRNAdb, Rfam, miRBase, starBase/ENCORI, and NONCODE, have been improved to support annotation and network analysis55,56,57,58,59.
Research on the roles of regulatory RNAs throughout the cell has accelerated. Studies incorporating advanced deep sequencing techniques have provided important insights through measurements of RNA structure, dynamics, and affinity. Guided by base pairing, regulatory RNAs have physiological functions in development, the cell cycle, and disease processes, influencing various biochemical pathways within the cell. A plausible complete sequence of each chromosome has only recently become accessible through long-read sequencing. Therefore, most of the functions of regulatory RNAs remain unclear60. In this review, we explore the current knowledge of regulatory RNAs, focusing on their structural dynamics and function in cellular components. We discuss nucleobase modification and protein cooperation in cellular mechanisms and, at the end of the review, we present RNA-based clinical approaches and ideas for using deep learning techniques.
RNA dynamics-based regulatory functions
The activities and functions of RNAs are influenced by their secondary or tertiary structures, which can be altered through various factors involving the binding of DNA, other RNAs, metal ions, proteins and metabolites, posttranscriptional modifications, mutations, and alterations in environmental conditions (Fig. 1). In recent years, multifaceted analytical approaches have been used to demonstrate correlations between RNA conformational dynamics and cellular functions, including the regulation of gene expression, alternative splicing, ribonucleoprotein assembly, and miRNA maturation. Thus, a comprehensive understanding of these structures can contribute to elucidating their functional mechanisms61,62.
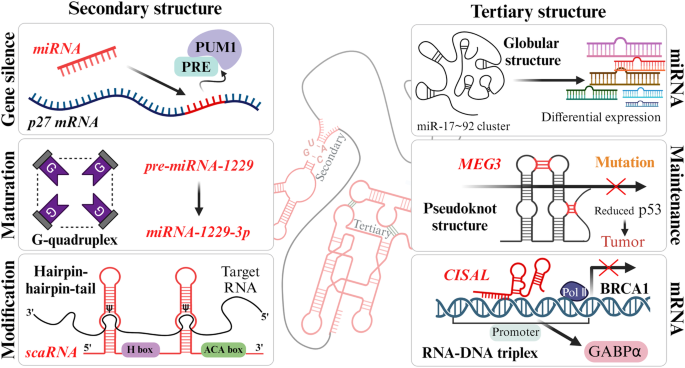
The dynamic structure of RNA is simply classified as the secondary a tertiary structures formed by specific base pairing and numerous factors, and various RNAs affect biological functions via these structures. In regulatory RNAs, the secondary structure is related to gene silencing via miRNA expression, miRNA maturation, and RNA modifications via scaRNAs. The tertiary structure participates in the regulation of p53 and the differential expression of miRNAs and mRNAs.
The secondary structure of an RNA is formed by interactions between specific nonadjacent bases in the primary nucleotide sequence. The four canonical RNA bases, adenine (A), uracil (U), guanine (G), and cytosine (C), are paired through hydrogen bonds according to the Watson-Crick principle (A-U and G-C), and wobble base pairing between G and U often occurs. These hydrogen bond-based base pairs contribute significantly to the stabilization and function of RNA and result in several secondary structure conformations, such as hairpin loops, bulge loops, inner loops, multibranched loops, single-stranded regions, helices and pseudoknots63,64,65. Gene silencing is a type of posttranscriptional process mediated by an RNA switch caused by miRNA binding. For instance, the 3â² UTR of p27 mRNA serves as the region where this RNA switch regulates mRNA expression. This involves the simultaneous release of the Pumilio-recognition element (PRE) from the Pumilio RNA-binding family member 1 (PUM1) cofactor, allowing miRNA binding. The interaction between PRE and PUM1 initiates the formation of the secondary structure that exposes the target site of the mRNA, promoting miRNA-mediated silencing66. Pre-miRNA-1229 is processed into mature miR-1229-3p through a noncanonical secondary structure, namely, a G-quadruplex that is balanced by canonical hairpin loops. In Alzheimerâs disease (AD), pre-miRNA-1229 containing the SNP rs2291418 induces structural malformations associated with pathological progression67. On the other hand, the scaRNA is the specific guide RNA for 2â²-O-methylation (2â²-O-Me) and pseudouridylation (Ï), which are associated with posttranscriptional modification of snRNA maturation in spliceosomal small nuclear RNPs (snRNPs). This RNA forms a âhairpinâhingeâhairpinâtailâ secondary structure with the target snRNA through conserved H/ACA box elements, contributing to Ï via specific factors68,69.
Tertiary structures are three-dimensional RNA building blocks resulting from the assembly of two or more secondary structures. These conformations are mediated via base stacking, noncanonical base pairing, the formation of triplex structures involving base triples, metal ions, metabolites, and interactions between unpaired bases and the riboseâphosphate backbone. The interactions between secondary structures and cofactors consequently lead to the formation of higher-order tertiary RNA structures with specific functions, such as scaffolding, regulation, and catalysis61,70. One cluster of pri-miRNAs contains six putative miRNA sequences: miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a. These six pri-miRNAs show different efficiencies for pre-miRNA generation in an RNA tertiary structure-dependent manner, as distal miRNAs are released more efficiently than internalized core proximal miRNAs in a globular tertiary structure. This three-dimensional conformation-specific expression of miRNAs from the miRNA cluster miR-17â92 is closely related to development and tumorigenesis71,72,73. Additionally, the human maternally expressed gene 3 (MEG3) lncRNA is composed of two highly conserved distal motifs, which are connected via complementary base pairing as a pseudoknot structure (kissing loops). MEG3 regulates the p53 pathway as a tumor suppressor, while mutated MEG3 inhibits the formation of the pseudoknot, leading to a reduction in the p53 response74. The lncRNA LINC01011, a cisplatin sensitivity-associated lncRNA (CISAL), regulates mitochondrial fission in tongue squamous cell carcinoma. CISAL forms an RNAâDNA triplex structure by binding directly to the promoter of breast cancer 1 (BRCA1), which induces the dissociation of GA-binding protein transcription factor subunit alpha (GABPα), the transcription factor for BRCA175.
Modification of RNA structures has consistently been implicated in numerous diseases, including heart failure and neurological disorders, such as AD. However, these dynamic conformations may form within picoseconds, while the formation of others may take seconds, with various factors modifying RNA structures intracellularly and extracellularly76,77,78. Therefore, further studies are needed to elucidate the background of RNA structure-specific cellular functions under multiple conditions.
Regulatory RNAs in cellular components
The versatile physiological functions of regulatory RNAs are dependent on their subcellular distribution (Fig. 2). Generally, ncRNAs are largely localized in subcellular compartments based on their molecular phenotypes, thus actively contributing to cellular homeostasis. Native transcripts of ncRNAs, after maturation, are located in the nucleus or cytoplasm, with a relatively greater percentage in the nucleus than in the cytoplasm79,80. Posttranscriptional regulation directs the proper localization of most transcripts via RNA cis-acting elements and their corresponding functions81.
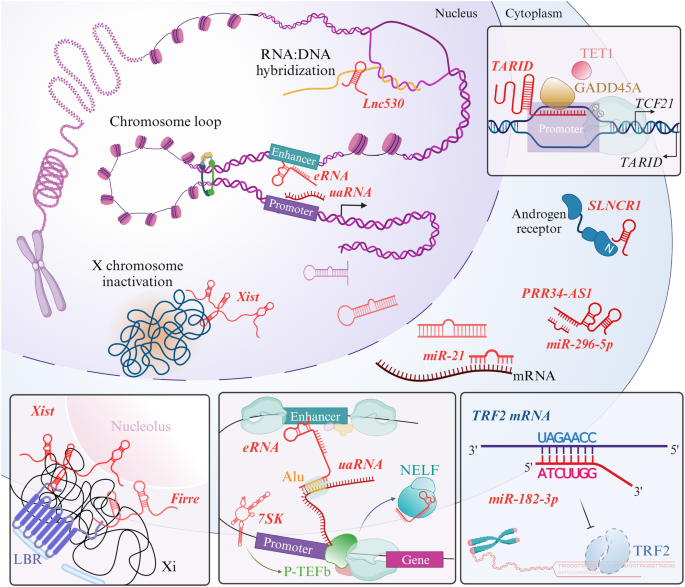
Physiological interactions of regulatory RNAs with the cellular machinery occur in both the nucleus and cytoplasm. In the nucleus, chromosome looping facilitates enhancer and promoter interactions to regulate gene expression; here, enriched eRNAs on active enhancer regions and uaRNAs near promoters lead to selective binding. In RNA:DNA hybridization, R-loops can be formed by mRNAs and ncRNAs to regulate gene expression. Inactivated X chromosomes are located in proximity to the nuclear envelope and nucleolus through interactions with Xist and Firre. Most commonly, miRNAs undergo structural maturation and then participate in silencing other RNA sequences. Some other regulatory RNAs modulate the expression of cytoplasmic proteins that play a role in signaling.
The nucleus is divided into subcompartments, including chromatin territories, the nucleolus, nuclear speckles, paraspeckles, and Cajal bodies, which are not membrane-enclosed. As a result, regulatory RNAs are largely located in the nucleus. Localization in these subnuclear compartments enables regulatory RNAs to participate in the machinery, acting in diverse aspects of cellular processes.
Chromatin has an intricate architecture and largely serves as the biological control tower for various cellular processes, including transcriptional regulation, DNA replication, and the cell cycle. In its protein-dense assembly within the genome structure, its tightly regulated hierarchical folding ensures spatiotemporally proper gene expression82. Among the myriad of cues that can result in chromatin alterations, regulatory RNAs are emerging as accessible mediators for interactive cooperation. These regulatory RNAs dynamically interact with genomic loci in response to the demands of numerous cellular processes. Chromatin-associated RNAs predominantly act at cis-regulatory sites to promote gene expression, as revealed by RNA-chromatin sequencing analysis, providing a global view of proximally driven interactions in cells and showing a propensity for an increased repertoire of chromatin-associated RNAs83,84. Although the precise mechanisms involving regulatory RNAs remain unclear, accumulating research indicates that many RNAs, especially lncRNAs, are involved in mechanisms of chromatin modulation. For instance, the long intergenic ncRNA HOXA transcript at the distal tip (HOTTIP) is generated from the 5â² end of the HOXA locus and has been suggested to regulate gene expression at distal sites. The HOTTIP RNA specifically interacts with chromatin via chromosomal looping. Suppression of HOTTIP RNA expression abolishes the dense occupation of the WDR5/MLL protein complex on the 5â² HOXA cluster, whereas properly transcribed HOTTIP facilitates the activation of transcription and H3K4 methylation via mutual interdependence on WDR585. The HOTTIP RNA also mediates chromatin remodeling, targeting neighboring genes, including HOXA9, FAS, and miR196b, with a confirmed relationship to acute myeloid leukemia86. Another HOX cluster-related long intergenic ncRNA, HOX transcript antisense RNA (HOTAIR), whose sequence is located specifically within the HOXC sequence on chromosome 12, is known to regulate the chromatin state87. The HOTAIR RNA epigenetically silences gene expression by interacting with the lysine (K)-specific demethylase 1 (LSD1)/PRC2 protein complex located at promoter regions, leading to H3K4 and H3K27 methylations. The chromatin-modifying complex is supported by HOTAIR RNA as a scaffold to support its recruitment to sites near the chromatin of target genes and is composed of EZH2, EED, SUZ12, and RbAp46/4888. Moreover, PRC2 was revealed to be dispensable for the repression of the expression of certain genes by the HOTAIR RNA89. Several X chromosome-embedded regulatory RNAs, such as X-inactivation specific transcript (Xist) and Firre intergenic repeating RNA element (Firre), which are lncRNAs that are X-linked and participate in maintaining nuclear organization, have been shown to be restricted to the nucleus. Xist is a conserved and well-characterized regulatory RNA in terms of X chromosome inactivation (Xi) that performs multiple intranuclear processes through chromosome-wide regulation. Xist triggers gene silencing by coating the X chromosome at proximal sites and subsequently represses gene expression. The localization of Xi is also strongly related to Xist-associated assembly. The concomitant direct interaction of lamin B receptor (LBR) recruits Xi to the nuclear periphery, where the adjacent nucleolus appears to facilitate higher-order Xi loop formation and maintain heterochromatin90,91,92,93. In addition, other regulatory RNAs that modulate Xist RNA expression have been identified. One antisense RNA termed Tsix is required for homeostatic Xist expression, which prevents aberrant transcriptional activity of the X chromosome; on the other hand, XistAR, Xert, Jpx, and Ftx have been demonstrated to promote Xist expression in cis92,94,95,96. Firre is anchored on the inactivated X chromosome, cooperatively organizing intranuclear positioning with Xist. The nucleolar association and H3K27me3 modification suggest the localizing ability of Firre, although the relevant mechanisms are not fully understood92,97. In the context of regulatory RNA involvement in immune cells, myeloid RNA regulator of Bim-induced death (Morrbid RNA, initially identified as Gm14005) has been suggested to regulate Bcl2lII (a proapoptotic gene) at the transcriptional level. Cytokine-mediated Morrbid RNA expression induces the suppression of Bcl2lII in cis of the bivalent promoter, regulating the repressive histone mark H3K27me3 and EZH2 methyltransferase activity98.
The transient hybridization of RNA:DNA results in the formation of R-loops, which displace single-stranded DNA, an essential structure for genome-wide regulation, particularly the regulation of chromatin dynamics and transcription. Enriched R-loops can be observed at active genes across the genome, posing a threat to a stable DNA topology99. Abnormally established and accumulated R-loop structures can lead to stalled transcription and replication100. In addition to nascent RNAs that form R-loops, other classes of RNAs (such as regulatory RNAs) form or utilize this structure for gene expression through epigenetic regulation101. In mouse embryonic stem cells (mESCs), the lncRNA Lnc530 was revealed to regulate R-loop formation to prevent excessive accumulation of two local proteins, TDP-43 and DDX5102. Another lncRNA, TARID, also in mESCs, binds to GADD45A, which is followed by the recruitment of DNA demethylases to promote TCF21 transcription by the formation of R-loops that occupy the TCF21 gene promoter103. In addition to lncRNA-mediated R-loop formation, R-loops themselves tether lncRNAs, recruiting several proteins with functions in regulating target genes.
The essential regions responsible for transcriptional regulation across noncoding regions of the genome, namely, enhancers and promoters, coordinate to drive the expression of specific genes104. In an already open chromatin template where histone octamers are removed, transcription factors that recognize a subset of factors to be incorporated can bind. Most promoters are located upstream of coding genes, while enhancers can be located up- or downstream of genes distally. Both adopt a cis configuration with respect to the genes they regulate. Enhancers can be located spatially proximal to promoters due to chromosomal looping and the accommodation of multiple proteins, including transcription factors, RNA polymerases, and architectural proteins that anchor and cooperate for regulatory signaling to induce gene expression105. In some cases, multiple discrete enhancers may contribute to the broad induction of a specific gene, such as c-fos, under temporal or tissue-specific control106.
Sequential control by enhancer regions due to insufficient activity of a general promoter determines whether the promoter is activated. Enhancer RNAs (eRNAs) are a distinct class of ncRNAs that are uni- or bidirectionally synthesized from active enhancers and are generally expressed at low levels107. These eRNAs were first identified in neurons and indeed were shown to regulate genes under conditions of neuronal stimulation in response to membrane depolarization108. Under these conditions, an activity-dependent induced eRNA is produced during the transcription process to sequester the negative elongation factor (NELF) complex from the cognate promoter, promoting gene expression. The eRNA specifically recognizes the RNA recognition motif (RRM) embedded in the NELF-E subunit, facilitating the release of RNA polymerase II pausing to resume elongation109 and stabilizing chromosomal looping via a cohesion complex28,110. The ability of eRNAâNELF binding to release NELF was further investigated and found to be dependent on the preference of NELF for guanosine nucleosides within sequences of more than 200ânt, which are bound by the multivalent tentacles of NELF111. The mechanism through which enhancers find their cognate promoters is largely unclear, recently, transposable sequence-mediated pairwise enhancer-promoter interactions throughout the genome have been suggested. Genome-wide mapping through RIC-seq has enabled us to understand the complementary interactions between RNAâRNA pairs and their potential mediator sequences. Enhancer-promoter selectivity has been suggested to be mediated by eRNA and antisense transcripts upstream of the promoter (i.e., uaRNAs), and 37.9% of the juxtaposing sequence in the eRNA-uaRNA interaction was found to be Alu elements112. Super-enhancer regions have been identified and assumed to determine cell identity via multiple clustered enhancers113,114. The eRNAs inferred to promote enhancer activity were repeatedly expressed at the highest levels when controlled by superenhancers, which may govern tumor heterogeneity115. With respect to the cooperation of enhancers and promoters, the 7SK snRNA is considered an essential regulator of transcription that forms a complex with positive transcription elongation factor b116. They were demonstrated to promote transcription in two ways, via mRNA or RNA polymerase II-specific snRNAs, and are involved in the repressive modulation of eRNAs117,118.
After nuclear export, mRNAs undergo translation, and the miRNA biogenesis pathway similarly progresses from the nucleus to the cytoplasm. Although miRNAs are more abundant in the cytoplasm than in the nucleus, nuclear-resident miRNAs have recently been recognized for their noncanonical roles119,120. Several lines of evidence suggest that miRNAs function in the nucleus through the nucleocytoplasmic transport of the RNA-RISC complex. The RISC cleaves transcripts, suggesting that miRNAs play a role in guiding this complex to cognate regions, even on RNAs of other classes121,122,123. Outside the nucleus, exported mRNA sequences undergo further processing for translation into proteins. miRNAs are known to regulate posttranscriptional gene expression through recognition of base pairs, particularly in the 3â²-UTRs of mRNAs in the cytoplasm. This event has historically been considered an important negative regulator of mRNA stability and translation18. The gene-silencing machinery directed by miRNAs can influence any mechanistic layer tuned for operation by the proper abundance of a protein. A nearly perfect complementary match (seed sequence) of 2â7 nucleotides targets a transcript, leading to its degradation and translational repression. Numerous types of miRNAs have been identified and revealed to be functionally involved in a myriad of cellular processes. For example, miR-10a-3p regulates KLF3-AS1 and ZBTB20 to promote apoptosis and extracellular matrix synthesis124, and miR-182-3p targets TRF2 to promote telomeric integrity125. One of the sophisticated miRNAs is miR-21, which targets PDCD4, TPM1, and PTEN for tumor suppression and is involved in diverse processes in cellular activities126,127. Notably, miRNAs can be sequestered by other competitive endogenous RNAs that share complementary sequences, ensuring the regulation of translation128. However, many roles of miRNAs in certain physiological processes have not yet been fully elucidated through identification of their cognate partners or cooperative molecules.
Cell signaling, as a fundamental communication ability of cells, is initiated and maintained by associated molecular cascades. Similarly, lncRNAs are crucial regulators in the cytoplasm, where they participate in intracellular cascades129,130. The lncRNA steroid receptor RNA activator-like noncoding RNA 1 (SLNCR1) interacts with the N-terminus of the androgen receptor to regulate downstream pathways. The SLNCR1 RNA occupies the androgen receptor in a sequence-dependent manner, activating a proinvasion signaling cascade in melanoma131. Cardiac apoptosis-related lncRNA (Carlr) preferentially interacts with p65 before its nuclear translocation, which regulates NF-κB signaling-associated genes132. PRR34 antisense RNA 1 (PRR34-AS1) was shown to regulate the Wnt/β-catenin pathway by sequestering miRNA-296-5p, which regulates E2F2 and SOX12133. The extracellular environment is another location where regulatory RNAs function. The dysfunctional telomere-derived shortened form of telomeric repeat-containing RNA was found to be secreted in exosomes, thus transmitting cytokine signals to other cells134. Beyond the cellular localization of regulatory RNAs, spatial transcripts across tissues also perform distinct functions according to the spatial context.
The highly discrete compartment-dependent distribution and constellation of regulatory RNAs need to be finely tuned to subsequently control the molecular machinery to maintain cellular homeostasis. Consequently, many undefined aspects of regulatory RNA-associated phenomena need to be delineated.
Regulatory RNAs involved in epitranscriptomic modifications
DNA and histone modifications are common factors mediating epigenetic regulation135,136,137. In recent research, posttranscriptional RNA modifications such as N6-methyladenosine (m6A), 5-methylcytosine (m5C), N1-methyladenosine (m1A), 2â²-O-Me, adenosine-to-inosine (A-to-I) editing, and Ï have been comprehensively investigated. These are known as epitranscriptomic regulators that play important roles in regulating gene expression and various cellular processes. The reciprocal impact of RNA modifications on regulatory RNAs underscores the intricate interplay between epitranscriptomic modifications and regulatory RNA-mediated processes138,139 (Fig. 3). Therefore, the importance of RNA modifications in epitranscriptomic regulation is increasingly recognized as a crucial aspect of cellular function140,141.
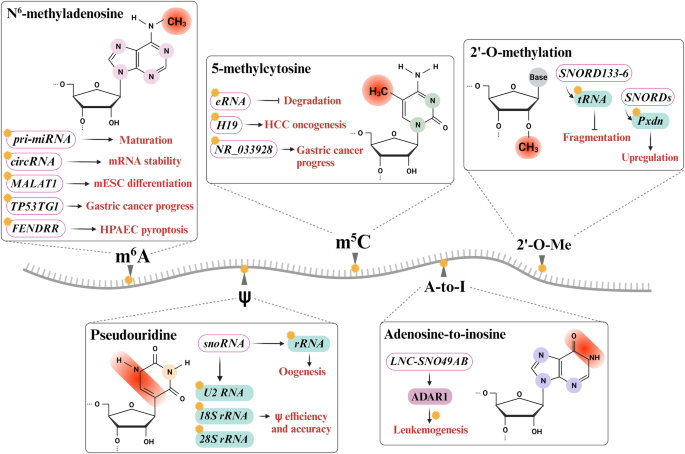
Many RNA modifications, including m6A, m5C, 2â²-O-Me, A-to-I, and Ï, have been identified in various RNAs. Multiple epitranscriptomic RNA modifications regulate and modulate cellular functions such as miRNA maturation, mRNA stability, gene expression, translation efficiency, degradation, and ESC differentiation. Additionally, these modifications are associated with various pathophysiological conditions, including gastric cancer, HCC, and leukemogenesis.
N6-methyladenosine (m6A)
m6A, methylation of the adenosine at the N6 position, is the most characterized and abundant internal RNA modification. This modification has been identified in coding and noncoding RNAs, such as mRNAs, tRNAs, rRNAs, lncRNAs, miRNAs and eRNAs. In particular, it is predominantly found in 5â²- and 3â²-UTRs, long internal exons, and near stop codons and is associated with the RRACH (Râ=âA or G, Hâ=âA, C, or U) consensus sequence. This modification is catalyzed by âwriterâ complexes, such as the methyltransferase 3 (METTL3), METTL5, METTL14, METTL16, Wilms tumor 1-associated protein, RNA binding motif 5, and zinc finger CCCH-like domain-containing protein 13 complexes. These m6A methyltransferase complexes participate in the process of target RNA modification142,143,144. The removal of m6A is orchestrated by several âerasersâ, including fat mass and obesity-associated protein and human AlkB homolog H5 (ALKBH5) with α-ketoglutarate. The âreadersâ of m6A, such as YTH domain-containing family (YTHDF), YTH domain-containing 2 and insulin-like growth factor 2 mRNA-binding protein 1, recognize m6A at different positions, thereby regulating biological functions involved in transcription, alternative splicing, RNA decay, RNA stability, nuclear export, and translation145,146,147. For instance, pri-mRNAs undergo several cleavage steps in the conversion from precursor miRNAs (pre-miRNAs) to mature miRNAs during miRNA biogenesis, which is regulated by various factors, including RNA methylation. Pri-miRNAs have enrichment of an m6A motif (GGAC), and it can undergo m6A modification by METTL3, which is recognized by heterogeneous nuclear ribonucleoprotein A2/B1 and causes miRNA processing via DiGeorge syndrome critical region 8. Thus, the level of a mature miRNA is modulated by the level of m6A deposited via METTL3148,149. Similar to mRNAs, circRNAs have numerous m6A modifications150. m6A-induced circRNAs participate in protein synthesis via the interaction of YTHDF3 and eIF4G2, which is an initiation factor. In addition, recognition of m6A-modified circRNAs by YTHDF2 is reported to be the key element in RNA degradation, and it also controls the stability of mRNAs and circRNA-associated immunity151,152. Previous studies have reported that m6A-modified lncRNAs regulate RNAâprotein and RNAâRNA interactions, such as the binding of heterogeneous nuclear ribonucleoprotein C to the U5 tract of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) lncRNA and the interactions of large intergenic coding RNA 1281 (linc1281) with the let-7 family of miRNAs, which are associated with mESC differentiation153,154. In addition, aberrant m6A regulation is significantly associated with diverse types of human diseases, including cancers, brain diseases, and cardiovascular diseases155,156,157. The level of TP53 target gene 1 (TP53TG1) lncRNA, a critical tumor suppressor, is decreased in gastric cancer. TP53TG1 has abundant m6A modification sites, but the demethylase ALKBH5 abolishes TP53TG1 stability, reduces its expression level and promotes its degradation, and suppresses the PI3K/AKT pathway by binding cellular inhibitor of protein phosphatase 2A, resulting in the inhibition of cell cycle progression and proliferation in gastric cancer. Thus, m6A-mediated TP53TG1 downregulation is closely related to the progression of gastric cancer158. The level of the lncRNA fetal-lethal noncoding developmental regulatory RNA (FENDRR) is markedly reduced in human pulmonary artery endothelial cells (HPAECs), which leads to hypoxia-induced HPAEC pyroptosis. The m6A âreaderâ YTHDC1 recognizes m6A-modified FENDRR and induces its degradation. m6A-modified FENDRR interacts with the dynamin-related protein 1 (DRP1) promoter by forming an RNAâDNA triplex that mediates its methylation and alters its transcription; therefore, DRP1 expression is low in healthy individuals. However, the m6A-mediated decay of FENDRR via interaction with YTHDC1 increases DRP1 expression, contributing to HPAEC pyroptosis159.
5-methylcytosine (m5C)
m5C, methylation of the fifth carbon of the RNA base in cytosine (C), is another common RNA modification. To date, m5C has been detected in a wide range of RNAs, including mRNAs, tRNAs, rRNAs, and eRNAs. Among these RNAs, m5C is abundant in eukaryotic tRNA and rRNA160,161. Members of the DNA methyltransferase-like 2 and NOL1/NOP2/SUN domain family member (NSUN) families mediate m5C modification in specific types of RNA. In addition, members of the ten-eleven translocation family function as m5C demethylases, while Aly/REF export factor and Y-box binding protein 1 act as readers to recognize m5C by binding to m5C-modified sites162,163. m5C modification contributes to multiple cellular functions that differ by RNA subtype140,163. m5C modification of eRNA prevents its degradation164. Moreover, m5C modification is closely related to the progression of various types of cancer160,165,166. The H19 lncRNA is aberrantly overexpressed and is known to have carcinogenic effects as a tumor-associated factor. In hepatocellular carcinoma (HCC), both the H19 lncRNA and m5C methylation are increased, which is significantly related to m5C-induced H19 lncRNA expression and is uniquely connected to the Ras-GTPase-activating protein SH3 domain-binding protein 1 (G3BP1). G3BP1, an oncoprotein, can bind to MYC mRNA, which causes its decay and promotes tumor progression. The increased level of m5C-modified H19 lncRNA can compete with G3BP1, which suppresses the degradation of MYC mRNA and induces an increase in the MYC level, thus enhancing the oncogenic process167. In gastric cancer, the lncRNA NR_033928 is upregulated and highly m5C methylated by overexpressed NSUN2, which contributes to poor prognosis. m5C-modified NR_033928 was found to promote an increase in glutaminase, which led to an increase in α-ketoglutarate (α-KG), a glutamine metabolite, and α-KG upregulated the expression of NR_033928 by activating 5-hydroxymethylcytosine demethylation, which promoted the progression of gastric cancer168.
2â²-O-methylation (2â²-O-Me)
2â²-O-Me, a highly conserved posttranscriptional modification of RNA, refers to the attachment of a methyl group to the 2â² hydroxyl (âOH) of the ribose moiety in any nucleotide via SNORDs and the 2â²-O-methyltransferase fibrillarin. This modification has been identified in a wide range of RNAs, including mRNAs, tRNAs, rRNAs, and snRNAs, which play important roles in various cellular processes. For instance, SNORDs and fibrillarin-mediated 2â²-O-Me in peroxidasin (Pxdn) promote the upregulation of Pxdn mRNA expression169. In the human ribosome, 2â²-O-methylation of rRNA can be regulated at functional sites important for translation, and the intrinsic capabilities of the ribosome modulate the translation of mRNA by altering the 2â²-O-Me pattern170. In addition, SNORD113-6-mediated 2â²-O-Me modification of tRNALer(TAA) inhibits the site-dependent fragmentation of a tRNA fragment (tRF)Leu, which is associated with vascular remodeling171. Moreover, 2â²-O-Me modification is correlated with lncRNA modulation. The ZFAS1 lncRNA and snoRNP NOP58 are upregulated in colorectal cancer (CRC), and ZFAS1 interacts with NOP58, which results in the binding of SNORD72C and SNORD78 to mediate 2â²-O-Me at the Gm3878 and Gm4593 sites in rRNA. As a result, the mRNA stability and translation of the downstream targets eukaryotic translation initiation factor 4A3 and laminin subunit gamma 2 increase, which promotes CRC cell proliferation and progression172. However, not all SNORDs do not participate in methylation. In HCC, lncRNA associated with liver regeneration (LALR1) and SNORD72 are highly upregulated, and LALR1 binds to SNORD72, which promotes the stability of inhibitor of DNA binding 2 mRNA. Subsequently, it enhances invasion and tumor growth173.
Adenosine-to-inosine (AâI) editing
A-to-I editing is the conversion of adenosine to inosine, via the deamination of the targeted adenosine to inosine. This reaction is catalyzed by adenosine deaminase acting on RNAs (ADARs), of which there are three, i.e., ADAR1, ADAR2, and ADAR3, but ADAR3 lacks catalytic activity174,175. The snoRNA-related lncRNA LNC-SNO49AB has two C/D box snoRNA sequences, SNORD49A and SNORD49B, which also contain a 5 m7G cap and a 3â² snoRNA structure. It is highly upregulated in leukemia and promotes not 2â²-O-Me modification but A-to-I editing by binding to ADAR1, which promotes hematopoietic malignancy176.
Pseudouridine (Ï)
Ï, the first confirmed RNA modification, identified in 1951, is an isomer of the nucleoside uridine that is composed of a carbonâcarbon (C1âC5) glycosidic bond instead of a nitrogen-carbon (C1-N1) glycosidic bond. Ï is significantly conserved across species and is found in most cellular RNAs, including mRNAs, tRNAs, rRNAs, and snRNAs. This modification is mediated by pseudouridine synthases, whose unique cellular localization and specific RNA targets have been reported177,178. Additionally, several unique targets need the participation of the RNAâprotein complex H/ACA box RNP, which is composed of four distinct proteins and a guide RNA and localized in the nucleolus and nucleoplasmic Cajal bodies. snoRNAs, as small nuclear guide RNAs, designate the regions of Ï modifications mediated by H/ACA box RNPs179,180. The intronic H/ACA snoRNA leads to pseudouridylation in the large rRNA subunit at positions 2258 and 2260 and is termed snR191 in yeast and hU19 in humans; the highly conserved Ï modification is beneficial for the cell181. Specifically, in oocytes, Ï modification of rRNA through the H/ACA box snRNP complex is required during oogenesis182. Furthermore, H/ACA box guide RNAs are composed of two hairpins that carry the internal Ï guide loops that allow site-dependent Ï of spliceosomal and ribosomal RNAs in humans. A recent study reported four human H/ACA RNAs (SNORA53, SNORA57, scaRNA8, and scaRNA1) that transfer Ï loops to promote the maintenance of two specific Ï modifications on U2 (Ψ43/Ψ44 and Ψ89/Ψ91) RNA, 18S (Ψ1045/Ψ1046), and 28S (Ψ3747/Ψ3749). These Ï loops have advantages in terms of efficiency and accuracy because of the versatility of Ï183.
Epitranscriptomic regulation via RNA modifications has been gradually investigated for its unique functions in various cellular processes and human diseases through advanced sequencing techniques. Novel RNA modifications in regulatory RNAs have continually been reported, and this evidence has contributed to the clarification of biological functions of RNAs. However, there are still numerous conflicting factors demonstrating the biochemical properties of RNAs inside and outside the cell; thus, additional studies are needed to determine the versatility of RNA modifications.
Interactions of regulatory RNAs with RNA-binding proteins
Regulatory RNAs themselves can be functional units; however, they mostly form dynamic RNAâprotein partnerships and function as central mediators or in mutual support. These interactions involve the binding of RNA to proteins and can occur reciprocally, with proteins recognizing RNA specifically through various RNA-binding domains such as the RRM, double-stranded RNA binding motif (dsRBM), DEAD/DEAH-box helicase domain, K homology (KH) domain, and zinc finger. Notably, recent studies have expanded our understanding of RNAâprotein interactions beyond conventional binding domain-mediated modules184. In this context, the functional bridge constructed by interactions between RNA and proteins plays intricate biological roles, contributing to the complexity of the cellular regulatory network (Fig. 4).
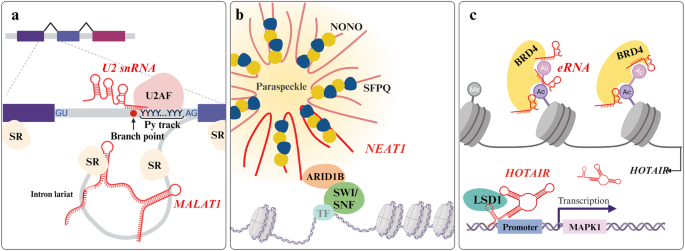
a U2 snRNA participates in the splicing process by recognizing the branch point, a process assisted by U2AF on the Py track near the 3â splice site. MALAT1 and SR proteins form intron lariats for alternative splicing. b NEAT1 is a well-defined scaffolding lncRNA localized in paraspeckles in the nucleus that can sequester proteins such as NONO and SFPQ. NEAT1 also mediates chromatin remodeling by binding to ARID1B. c BRD4 recognizes acetylated K residues in a manner mediated by eRNAs and regulates HOTAIR expression levels. HOTAIR facilitates transcriptional regulation via LSD1 recruitment.
Prior to the nuclear export of synthesized RNA transcripts, primary transcripts undergo an RNA processing program that is elaborately orchestrated by numerous macromolecules. This processing program includes splicing and maturation, resulting in the establishment of a highly diversified transcriptome and proteome. A prominent example of higher-order assembly is the spliceosome, a multimegadalton macromolecular apparatus that participates in and regulates splicing tightly in direct and indirect ways and involves the participation of regulatory RNAs185. Nuclear speckles, located in the interchromatin space, provide platforms for preparing the splicing of diverse classes of RNAs186. The representative splicing process begins when the spliceosome binds to the splice sites of heterogeneous nuclear RNAs (hnRNAs), forming a complex referred to as an snRNP that is composed of snRNAs and confers recognition ability and catalytic activity. This module mediates transesterification and sequence rearrangement187. The U2 snRNA is a major snRNA that facilitates the recognition of the intronic branch point via nucleotide base pairing, initiating recognition of the 3â² splice site. The recognition of the 3â² splice site is facilitated by U2 auxiliary factor (U2AF) due to branch point degeneracy188,189. The heterodimeric U2AF complex (containing 65 and 35âkDa subunits) specifically detects the polypyrimidine (Py) tract element upstream of the 3â² splice site and then accomplishes spliceosome assembly followed by lariat formation for the excision of the designated intron sequence190. Nuclear speckles for the assembly of the large spliceosome are constructed via the contribution of lncRNAs. MALAT1, a highly expressed lncRNA, has been demonstrated to recruit serine- and arginine-rich (SR) proteins that dock to pre-mRNA sequences, thereby regulating splicing191. MALAT1 not only interacts with SR splicing factors but also governs phosphorylation levels, subsequently influencing alternative splicing events192. Two splicing factor proteins, Py tract-binding protein 1 and PTB-associated splicing factor (PSF), are implicated as other regulatory coordinators in splicing, interacting with MALAT1. PTBP1 and PSF become stabilized through direct interaction with MALAT1, resulting in the exclusion or inclusion of exons during alternative splicing193. Splicing and maturation also occur in other classes of RNAs through component regulatory RNAs and RNPs: in ribosomal RNAs in the nucleolus via snoRNAs, in snRNAs in Cajal bodies via scaRNAs, and in histone pre-mRNA in histone locus bodies via snRNAs during processing194. These regions are transcriptionally active and are referred to as membraneless nuclear bodies.
As nuclear bodies, paraspeckles are sphere-like RNP granule structures that utilize NEAT1 lncRNA as an assembly scaffold. These structures are commonly found in proximity to nuclear speckles. Although substantial information is needed for a full understanding, paraspeckles are suggested to function as platforms that segregate certain RNAs or proteins, resulting in molecular functions associated with the sequestered molecules195. The splicing factor proline/glutamine-rich (SFPQ) plays versatile roles by acting as a splicing factor and transcriptional regulator and is also a core component in paraspeckle retention through direct interaction with NEAT1. Increased paraspeckle formation is triggered under stressful conditions, such as intracellular invasion. This may sequester SFPQ, which is a suppressor of interleukin-8 cytokine transcription196 and a transcription factor of the ADARB2 gene197. Non-POU domain-containing octamer-binding protein (NONO) is another essential component that contributes to the coalescence of paraspeckles and regulates gene expression along with SFPQ. Additionally, NONO participates in the DNA double-strand break repair mechanism through recruitment of another protein, RPLP0. X-irradiation-induced DNA damage promotes NONO condensation in NEAT1-associated paraspeckles, and NONO then interacts with RPLP0, leading to nonhomologous end joining repair198. Moreover, recent findings have revealed the direct implication of NEAT1 in interacting with paraspeckles and the chromatin remodeling complex. ATP hydrolysis allows the repositioning of nucleosomes via binding to the switch/sucrose nonfermentable (SWI/SNF) complex, which increases the accessibility of other molecules for transcriptional regulation199. Coordination between SWI/SNF and paraspeckles by a specific interactor, ARID1B, has been demonstrated, and this interaction allows the recruitment of various kinds of chromatin-associated proteins to influence gene regulatory processes, leading to alternative splicing. This interaction was markedly detected only with the canonical BAF (cBAF) among the three distinct mammalian SWI/SNF complexes, and cBAF is known to govern the enhancer region200,201.
The nucleosome positioning process, which involves sliding or dissociating DNA for gene regulatory mechanisms, is typically referred to as chromatin remodeling. The flexibility of chromatin arises from its rearrangement through an ATP-dependent mechanism to open DNA grooves. The highly conserved bromodomain (BRD) recognizes acetylations at K residues on histone tails and induces the assembly of BRD-containing proteins primarily for transcriptional regulation202. The BRD4 protein contributes to super-enhancer organization, with tandem BRDs utilizing the eRNA-BRD4 interaction to promote tethering to acetylated K residues203. Alongside the conserved transcriptional regulatory elements for lncRNAs, BRD4 was found to have a direct interaction with the HOTAIR promoter, controlling the HOTAIR transcript level204. Regarding the role of HOTAIR after BRD4-mediated promotion, HOTAIR can attract LSD1 to decrease the H3K9me2 level of the MAPK1 promoter205. Several related lncRNAs, namely, H19, HOTAIRM1, DGCR5, and MEG3, were also revealed to be related to BRD4-associated regulation204. Furthermore, BRD4 plays a functional role after forming a complex with NEAT1, which is negatively regulated via alterations in WDR5 or EZH2 activity206. A recently identified lncRNA, LENGA, acts as a connector, enhancing the interaction between the transcription factor TP53 and BRD7. BRD7, a subunit of the polybromo-associated BAF SWI/SNF complex, uses regulatory RNAs as a scaffold to regulate downstream gene expression207. The core subunit SMARCB1 of SWI/SNF and its suggested partner, SMARCB1 lncRNA, together modulate the expression of the GAS6 oncogene208. The catalytic subunit BRG1 of the SWI/SNF complex interacts with the uc.291 lncRNA derived from ACTL6A to regulate the transcription of EDC genes209. As described above, based on these remarkably sophisticated intracellular contributions of regulatory RNAs, intercellular communication and a series of changes can be explained by the interdependence of proteins and regulatory RNAs.
Conclusions
In the rapidly evolving field of regulatory RNAs, novel and functional RNAs have been consistently uncovered, paving the way for the development of meaningful RNA-centric technologies based on modern biology. Notably, recent global efforts to combat coronavirus disease 2019 have accelerated the development of mRNA vaccines, which have shown remarkable potential as therapeutic countermeasures210,211,212. Within this context, various RNA-based therapeutics, including small interfering RNAs, antisense oligonucleotides (ASOs), CRISPR/Cas9 gene editing, anti-miRs (or antagomirs), aptamers, miRNA sponges, therapeutic circRNAs and mRNA vaccines, have emerged to address diverse pathologies. These therapeutics modulate the expression of mRNAs as well as ncRNAs by targeting cognate sequences213,214 and have been focused primarily on well-defined mRNA targets215,216,217,218,219. For instance, ASOs have been employed in the treatment of Duchenne muscular dystrophy, with casimersen gaining approval for its modulatory effects on splicing in intergenic regions220. Antagomirs, antisense miRNAs, exhibit nuclease resistance through 2â²-methoxy substitution. The results of clinical trials of MRG-110, targeting miR-92a, in angiogenesis and CDR132L, targeting miR-132, in heart failure (phase I: NCT03603431 and phase Ib: NCT03494712) underscore the therapeutic potential of antagomirs221. RNAs have a strong negative charge, which renders them plasma membrane-impermeable and susceptible to degradation by RNases, and thus their ability to attain physical proximity to their precise target sequences is limited, and efforts to overcome these challenges are ongoing219. Numerous studies are underway to overcome these limitations by exploring modifications to the phosphodiester backbone and employing viral-vector-based and nonviral delivery systems220,222, nanocarriers such as lipid nanoparticles, cationic polymer-based polyplexes, spherical nucleic acids, DNA nanostructures and exosomes219. For instance, the GalNAc-conjugated antagomir AZD4076 (RG-125), designed to improve the delivery of targeting ligands, was developed to target intracellular miR-103/107 but was discontinued in phase I trials (NCT02612662 and NCT02826525)214. Antagomirs, on the other hand, are strategically designed to exhibit increased permeability via conjugation to cholesterol223.
The continuous investigation of applicable regulatory RNAs and their effectiveness of delivery remains crucial for understanding their biological roles in response to a wide range of cellular and environmental conditions. As the field progresses, the exploration of innovative therapeutic approaches and delivery strategies holds promise for addressing the complex challenges associated with RNA-based interventions.
Many studies have attempted to predict the biological functions of regulatory RNAs, recognizing their substantial potential as key cellular regulatory factors224,225,226. However, defining only single time point-dependent conformations and versatilities of important RNAs is insufficient, as intrinsic and extrinsic factors differ between physiological and disease conditions. The important biochemical properties of RNAs, including their folding stability, binding affinity, specificity, modification sites, and catalytic efficiency, still cannot be predicted by sequencing-based quantitative measurements61,138,227,228. Undoubtedly, deep learning approaches have become important for expanding strategic analyses to apply RNA-related predictions using various sequencing-based datasets229,230,231,232,233. The data-driven approach excels at extracting optimal patterns and knowledge through sophisticated computational processes, leveraging massive training data and convolution operations in multiple hidden layers within various neural network algorithms234,235,236. Recent studies have reported a wide range of RNA-related predictions using diverse deep learning methods, such as multiple types RNA modification sites predictor, ensemble multiscale deep learning predictor, Deepm5C, PseUdeep, Deep-2â²-O-Me, and DLm6Am for predicting modifications234,237,238,239,240,241; DeepMirTar, deep donor splice site recognizer, and RBPsuite for predicting specific target sites242,243,244; DeepLncLoc for predicting localization245; ncRDense and circDeep for predicting classification246,247; UFold, NuFold, and deep learning-ALIgned nucleic acids for predicting structure248,249,250; and rBPDL and DeepBtoD for predicting RNA-binding proteins251,252 (Table 2). However, the practical correlations among these results needs to be demonstrated for more accurate predictions because in deep learning approaches, information is acquired from the input data; therefore, batch variability may influence the output data. The accuracy of predictive analyses with genomic data-based deep learning seems to be significantly dependent on the quality and quantity of the training data. Therefore, deep learning requires more refined and integrated training data obtained from various studies. Therefore, precise and highly efficient results that previously could not be obtained via biochemical experiments can now be obtained. Accelerated research in this field employing cutting-edge techniques is expected to expand to establish strategic approaches for RNA-targeting therapies based on a comprehensive understanding of RNA regulatory mechanisms.
Owing to exponential advancements in modern biological techniques, we now have access to massive amounts of genome-wide information. Dissecting molecular layers, particularly within the RNA-associated cellular machinery, seems likely to provide a more comprehensive understanding. However, there are many hurdles to overcome in interpreting the functions of the myriad of regulatory RNAs, in accordance with their associated macromolecule complexes and distributions in both the nucleus and the cytoplasm. Regulatory RNAs are more prevalent than previously appreciated but exhibit lower sequence conservation and expression levels than coding RNAs. Addressing these challenges requires enormous effort and critical considerations, including exceeding the baseline of transcriptional noise due to technical limitations. Revealing the isoforms and stoichiometries of regulatory RNAs under various physiological conditions is crucial for obtaining a deeper understanding of these RNAs. Gaining such an understanding of the extensive properties of regulatory RNAs is expected to significantly increase insights into the intricacies of organisms.
References
Bizuayehu, T. T. et al. Long-read single-molecule RNA structure sequencing using nanopore. Nucleic Acids Res. 50, e120 (2022).
Guh, C. Y., Hsieh, Y. H. & Chu, H. P. Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J. Biomed. Sci. 27, 44 (2020).
Jones, C. P. & Ferre-DâAmare, A. R. RNA quaternary structure and global symmetry. Trends Biochem. Sci. 40, 211â220 (2015).
Chen, K. et al. LncRNA SNHG6 promotes glycolysis reprogramming in hepatocellular carcinoma by stabilizing the BOP1 protein. Anim. Cells Syst. 26, 369â379 (2022).
Wright, B. W., Yi, Z., Weissman, J. S. & Chen, J. The dark proteome: translation from noncanonical open reading frames. Trends Cell Biol. 32, 243â258 (2022).
Huang, Z., Zhou, J. K., Peng, Y., He, W. & Huang, C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 19, 77 (2020).
Kaikkonen, M. U., Lam, M. T. & Glass, C. K. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 90, 430â440 (2011).
Wei, L. et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol. Cancer 18, 147 (2019).
Morris, K. V. & Mattick, J. S. The rise of regulatory RNA. Nat. Rev. Genet. 15, 423â437 (2014).
Ransohoff, J. D., Wei, Y. & Khavari, P. A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143â157 (2018).
Yang, S., Lim, K. H., Kim, S. H. & Joo, J. Y. Molecular landscape of long noncoding RNAs in brain disorders. Mol. Psychiatry 26, 1060â1074 (2021).
Mattick, J. S. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430â447 (2023).
Statello, L., Guo, C. J., Chen, L. L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96â118 (2021).
Lim, K. H., Yang, S., Kim, S. H., Chun, S. & Joo, J. Y. Discoveries for long non-coding RNA dynamics in traumatic brain injury. Biology 9, https://doi.org/10.3390/biology9120458 (2020).
Kim, S. H., Lim, K. H., Yang, S. & Joo, J. Y. Long non-coding RNAs in brain tumors: roles and potential as therapeutic targets. J. Hematol. Oncol. 14, 77 (2021).
Sartorelli, V. & Lauberth, S. M. Enhancer RNAs are an important regulatory layer of the epigenome. Nat. Struct. Mol. Biol. 27, 521â528 (2020).
Han, Z. & Li, W. Enhancer RNA: what we know and what we can achieve. Cell Prolif. 55, e13202 (2022).
Shang, R., Lee, S., Senavirathne, G. & Lai, E. C. microRNAs in action: biogenesis, function and regulation. Nat. Rev. Genet. 24, 816â833 (2023).
Khanbabaei, H. et al. Non-coding RNAs and epithelial mesenchymal transition in cancer: molecular mechanisms and clinical implications. J. Exp. Clin. Cancer Res. 41, 278 (2022).
OâBrien, J., Hayder, H., Zayed, Y. & Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 9, 402 (2018).
Ng, K. W. et al. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol. Cancer 15, 5 (2016).
Cao, T. et al. Biology and clinical relevance of noncoding sno/scaRNAs. Trends Cardiovasc. Med. 28, 81â90 (2018).
Huang, Z. H., Du, Y. P., Wen, J. T., Lu, B. F. & Zhao, Y. snoRNAs: functions and mechanisms in biological processes, and roles in tumor pathophysiology. Cell Death Discov. 8, 259 (2022).
He, A. T., Liu, J., Li, F. & Yang, B. B. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal. Transduct. Target Ther. 6, 185 (2021).
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675â691 (2019).
Sun, Y. M. & Chen, Y. Q. Principles and innovative technologies for decrypting noncoding RNAs: from discovery and functional prediction to clinical application. J. Hematol. Oncol. 13, 109 (2020).
Hou, T. Y. & Kraus, W. L. Spirits in the material world: enhancer RNAs in transcriptional regulation. Trends Biochem. Sci. 46, 138â153 (2021).
Li, Q. et al. Enhancer RNAs: mechanisms in transcriptional regulation and functions in diseases. Cell Commun. Signal 21, 191 (2023).
Li, J., Batcha, A. M., Gruning, B. & Mansmann, U. R. An NGS workflow blueprint for DNA sequencing data and its application in individualized molecular oncology. Cancer Inf. 14, 87â107 (2015).
Hess, J. F. et al. Library preparation for next generation sequencing: a review of automation strategies. Biotechnol. Adv. 41, 107537 (2020).
Mumbach, M. R. et al. HiChIRP reveals RNA-associated chromosome conformation. Nat. Methods 16, 489â492 (2019).
Jung, N. & Kim, T. K. Advances in higher-order chromatin architecture: the move towards 4D genome. BMB Rep. 54, 233â245 (2021).
Bell, J. C. et al. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife 7, https://doi.org/10.7554/eLife.27024 (2018).
Bonetti, A. et al. RADICL-seq identifies general and cell type-specific principles of genome-wide RNA-chromatin interactions. Nat. Commun. 11, 1018 (2020).
Horlacher, M. et al. Towards in silico CLIP-seq: predicting protein-RNA interaction via sequence-to-signal learning. Genome Biol. 24, 180 (2023).
Jensen, K. B. & Darnell, R. B. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol. Biol. 488, 85â98 (2008).
Spitzer, J. et al. PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation): a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol. 539, 113â161 (2014).
Garzia, A., Meyer, C., Morozov, P., Sajek, M. & Tuschl, T. Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods 118-119, 24â40 (2017).
Konig, J. et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 17, 909â915 (2010).
Huppertz, I. et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65, 274â287 (2014).
Cai, Z. et al. RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature 582, 432â437 (2020).
Margasyuk, S. et al. RNA in situ conformation sequencing reveals novel long-range RNA structures with impact on splicing. RNA 29, 1423â1436 (2023).
Nguyen, T. C. et al. Mapping RNA-RNA interactome and RNA structure in vivo by MARIO. Nat. Commun. 7, 12023 (2016).
Zubradt, M. et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75â82 (2017).
Jin, Q., Zhang, L., Hu, S., Wei, G. & Wang, Z. Probing in vivo RNA structure with optimized DMS-MaPseq in rice. Front. Plant Sci. 13, 869267 (2022).
Morandi, E. et al. Genome-scale deconvolution of RNA structure ensembles. Nat. Methods 18, 249â252 (2021).
Sanz, L. A. & Chedin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 14, 1734â1755 (2019).
Halasz, L. et al. RNA-DNA hybrid (R-loop) immunoprecipitation mapping: an analytical workflow to evaluate inherent biases. Genome Res. 27, 1063â1073 (2017).
Wahba, L., Costantino, L., Tan, F. J., Zimmer, A. & Koshland, D. S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30, 1327â1338 (2016).
Sanz, L. A. et al. Prevalent, dynamic, and conserved r-loop structures associate with specific epigenomic signatures in mammals. Mol. Cell 63, 167â178 (2016).
Dumelie, J. G. & Jaffrey, S. R. Defining the location of promoter-associated R-loops at near-nucleotide resolution using bisDRIP-seq. Elife 6, https://doi.org/10.7554/eLife.28306 (2017).
Williams, C. G., Lee, H. J., Asatsuma, T., Vento-Tormo, R. & Haque, A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 14, 68 (2022).
Marx, V. Method of the year: spatially resolved transcriptomics. Nat. Methods 18, 9â14 (2021).
Riba, A. et al. Cell cycle gene regulation dynamics revealed by RNA velocity and deep-learning. Nat. Commun. 13, 2865 (2022).
Kozomara, A., Birgaoanu, M. & Griffiths-Jones, S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155âD162 (2019).
Zhao, L. et al. NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 49, D165âD171 (2021).
Kariuki, D. et al. Review of databases for experimentally validated human microRNA-mRNA interactions. Database 2023, https://doi.org/10.1093/database/baad014 (2023).
Kalvari, I. et al. Non-coding RNA analysis using the rfam database. Curr. Protoc. Bioinform. 62, e51 (2018).
Amaral, P. P., Clark, M. B., Gascoigne, D. K., Dinger, M. E. & Mattick, J. S. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 39, D146â151, (2011).
Lagarde, J. et al. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat. Genet. 49, 1731â1740 (2017).
Ganser, L. R., Kelly, M. L., Herschlag, D. & Al-Hashimi, H. M. The roles of structural dynamics in the cellular functions of RNAs. Nat. Rev. Mol. Cell Biol. 20, 474â489 (2019).
Vicens, Q. & Kieft, J. S. Thoughts on how to think (and talk) about RNA structure. Proc. Natl Acad. Sci. USA 119, e2112677119 (2022).
Lu, W. et al. Research on RNA secondary structure predicting via bidirectional recurrent neural network. BMC Bioinform. 22, 431 (2021).
Zhao, Q. et al. Review of machine learning methods for RNA secondary structure prediction. PLoS Comput. Biol. 17, e1009291 (2021).
Wang, X. W., Liu, C. X., Chen, L. L. & Zhang, Q. C. RNA structure probing uncovers RNA structure-dependent biological functions. Nat. Chem. Biol. 17, 755â766 (2021).
Kedde, M. et al. A pumilio-induced RNA structure switch in p27-3â UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 12, 1014â1020 (2010).
Imperatore, J. A., Then, M. L., McDougal, K. B. & Mihailescu, M. R. Characterization of a G-quadruplex structure in pre-miRNA-1229 and in its Alzheimerâs disease-associated variant rs2291418: implications for miRNA-1229 maturation. Int. J. Mol. Sci. 21, https://doi.org/10.3390/ijms21030767 (2020).
Richard, P. et al. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 22, 4283â4293 (2003).
Cerneckis, J., Cui, Q., He, C., Yi, C. & Shi, Y. Decoding pseudouridine: an emerging target for therapeutic development. Trends Pharm. Sci. 43, 522â535 (2022).
Dethoff, E. A., Chugh, J., Mustoe, A. M. & Al-Hashimi, H. M. Functional complexity and regulation through RNA dynamics. Nature 482, 322â330 (2012).
Chaulk, S. G. et al. Role of pri-miRNA tertiary structure in miR-17~92 miRNA biogenesis. RNA Biol. 8, 1105â1114 (2011).
Chakraborty, S., Mehtab, S., Patwardhan, A. & Krishnan, Y. Pri-miR-17-92a transcript folds into a tertiary structure and autoregulates its processing. RNA 18, 1014â1028 (2012).
Gan, H. H. & Gunsalus, K. C. The role of tertiary structure in MicroRNA target recognition. Methods Mol. Biol. 1970, 43â64 (2019).
Uroda, T. et al. Conserved pseudoknots in lncRNA MEG3 are essential for stimulation of the p53 pathway. Mol. Cell 75, 982â995.e989 (2019).
Fan, S. et al. lncRNA CISAL inhibits BRCA1 transcription by forming a tertiary structure at its promoter. iScience 23, 100835 (2020).
Bernat, V. & Disney, M. D. RNA structures as mediators of neurological diseases and as drug targets. Neuron 87, 28â46 (2015).
Rybak-Wolf, A. & Plass, M. RNA dynamics in Alzheimerâs disease. Molecules 26, https://doi.org/10.3390/molecules26175113 (2021).
Liu, W., Higashikuni, Y. & Sata, M. Linking RNA dynamics to heart disease: the lncRNA/miRNA/mRNA axis in myocardial ischemia-reperfusion injury. Hypertens. Res. 45, 1067â1069 (2022).
Carlevaro-Fita, J. & Johnson, R. Global positioning system: understanding long noncoding RNAs through subcellular localization. Mol. Cell 73, 869â883 (2019).
Mas-Ponte, D. et al. LncATLAS database for subcellular localization of long noncoding RNAs. RNA 23, 1080â1087 (2017).
Tong, C. & Yin, Y. Localization of RNAs in the nucleus: cis- and trans-regulation. RNA Biol. 18, 2073â2086 (2021).
Deng, S., Feng, Y. & Pauklin, S. 3D chromatin architecture and transcription regulation in cancer. J. Hematol. Oncol. 15, 49 (2022).
Limouse, C. et al. Global mapping of RNA-chromatin contacts reveals a proximity-dominated connectivity model for ncRNA-gene interactions. Nat. Commun. 14, 6073 (2023).
Li, X. et al. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 35, 940â950 (2017).
Wang, K. C. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120â124 (2011).
Singh, A. P. et al. A coordinated function of lncRNA HOTTIP and miRNA-196b underpinning leukemogenesis by targeting FAS signaling. Oncogene 41, 718â731 (2022).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071â1076 (2010).
Mozdarani, H., Ezzatizadeh, V. & Rahbar Parvaneh, R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J. Transl. Med. 18, 152 (2020).
Portoso, M. et al. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 36, 981â994 (2017).
Chen, C. K. et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468â472 (2016).
Brockdorff, N., Bowness, J. S. & Wei, G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 34, 733â744 (2020).
Dossin, F. & Heard, E. The molecular and nuclear dynamics of X-chromosome inactivation. Cold Spring Harb. Perspect. Biol. 14, https://doi.org/10.1101/cshperspect.a040196 (2022).
Engreitz, J. M., Ollikainen, N. & Guttman, M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756â770 (2016).
Sarkar, M. K. et al. An Xist-activating antisense RNA required for X-chromosome inactivation. Nat. Commun. 6, 8564 (2015).
Gjaltema, R. A. F. et al. Distal and proximal cis-regulatory elements sense X chromosome dosage and developmental state at the Xist locus. Mol. Cell 82, 190â208 e117 (2022).
Sun, S. et al. Jpx RNA activates Xist by evicting CTCF. Cell 153, 1537â1551 (2013).
Yang, F. et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16, 52 (2015).
Kotzin, J. J. et al. The long non-coding RNA morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537, 239â243 (2016).
Petermann, E., Lan, L. & Zou, L. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat. Rev. Mol. Cell Biol. 23, 521â540 (2022).
Skourti-Stathaki, K. & Proudfoot, N. J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 28, 1384â1396 (2014).
Boque-Sastre, R. et al. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl Acad. Sci. USA 112, 5785â5790 (2015).
Gong, D. et al. Long noncoding RNA Lnc530 localizes on R-loops and regulates R-loop formation and genomic stability in mouse embryonic stem cells. Stem Cell Rep. 18, 952â968 (2023).
Arab, K. et al. GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat. Genet. 51, 217â223 (2019).
Shlyueva, D., Stampfel, G. & Stark, A. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 15, 272â286 (2014).
Andersson, R. & Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71â87 (2020).
Joo, J. Y., Schaukowitch, K., Farbiak, L., Kilaru, G. & Kim, T. K. Stimulus-specific combinatorial functionality of neuronal c-fos enhancers. Nat. Neurosci. 19, 75â83 (2016).
Mikhaylichenko, O. et al. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 32, 42â57 (2018).
Kim, T. K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182â187 (2010).
Schaukowitch, K. et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell 56, 29â42 (2014).
Tsai, P. F. et al. A muscle-specific enhancer RNA mediates cohesin recruitment and regulates transcription in trans. Mol. Cell 71, 129â141.e128 (2018).
Gorbovytska, V. et al. Enhancer RNAs stimulate Pol II pause release by harnessing multivalent interactions to NELF. Nat. Commun. 13, 2429 (2022).
Liang, L. et al. Complementary Alu sequences mediate enhancer-promoter selectivity. Nature 619, 868â875 (2023).
Pott, S. & Lieb, J. D. What are super-enhancers? Nat. Genet. 47, 8â12 (2015).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934â947 (2013).
Chen, H. & Liang, H. A high-resolution map of human enhancer RNA loci characterizes super-enhancer activities in cancer. Cancer Cell 38, 701â715.e705 (2020).
Fujinaga, K., Huang, F. & Peterlin, B. M. P-TEFb: the master regulator of transcription elongation. Mol. Cell 83, 393â403 (2023).
Flynn, R. A. et al. 7SK-BAF axis controls pervasive transcription at enhancers. Nat. Struct. Mol. Biol. 23, 231â238 (2016).
Egloff, S. et al. The 7SK snRNP associates with the little elongation complex to promote snRNA gene expression. EMBO J. 36, 934â948 (2017).
Roberts, T. C. The microRNA biology of the mammalian nucleus. Mol. Ther. Nucleic Acids 3, e188 (2014).
Kim, J. Y., Kim, W. & Lee, K.-H. The role of microRNAs in the molecular link between circadian rhythm and autism spectrum disorder. Anim. Cells Syst. 27, 38â52 (2023).
Leucci, E. et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 3, 2535 (2013).
Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414â4422 (2011).
Laitinen, P. et al. Nuclear microRNA-466c regulates Vegfa expression in response to hypoxia. PLoS One 17, e0265948 (2022).
Chen, S. et al. The long non-coding RNA KLF3-AS1/miR-10a-3p/ZBTB20 axis improves the degenerative changes in human nucleus pulposus cells. Cell Tissue Res. 393, 97â109 (2023).
Dinami, R. et al. MiR-182-3p targets TRF2 and impairs tumor growth of triple-negative breast cancer. EMBO Mol. Med. 15, e16033 (2023).
Hill, M. & Tran, N. Global miRNA to miRNA Interactions: Impacts for miR-21. Trends Cell Biol. 31, 3â5 (2021).
Zhu, S. et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 18, 350â359 (2008).
Kartha, R. V. & Subramanian, S. Competing endogenous RNAs (ceRNAs): new entrants to the intricacies of gene regulation. Front. Genet. 5, 8 (2014).
Xu, S., Gong, Y., Yin, Y., Xing, H. & Zhang, N. The multiple function of long noncoding RNAs in osteosarcoma progression, drug resistance and prognosis. Biomed. Pharmacother. 127, 110141 (2020).
Jang, W., Im, M., Roh, J., Kang, J. & Kim, W. Hippo-YAP/TAZ pathway regulation: the crucial roles of lncRNAs in cancer. Anim. Cells Syst. 27, 309â320 (2023).
Schmidt, K. et al. Targeting the oncogenic long non-coding RNA SLNCR1 by blocking its sequence-specific binding to the androgen receptor. Cell Rep. 30, 541â554.e545 (2020).
Castellanos-Rubio, A. et al. Cytoplasmic form of Carlr lncRNA facilitates inflammatory gene expression upon NF-kappaB activation. J. Immunol. 199, 581â588 (2017).
Qin, M. et al. lncRNA PRR34-AS1 promotes HCC development via modulating Wnt/beta-catenin pathway by absorbing miR-296-5p and upregulating E2F2 and SOX12. Mol. Ther. Nucleic Acids 25, 37â52 (2021).
Wang, Z. et al. Telomeric repeat-containing RNA (TERRA) constitutes a nucleoprotein component of extracellular inflammatory exosomes. Proc. Natl Acad. Sci. USA 112, E6293âE6300 (2015).
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245â254 (2003).
Baxter, E., Windloch, K., Gannon, F. & Lee, J. S. Epigenetic regulation in cancer progression. Cell Biosci. 4, 45 (2014).
Zhang, L., Lu, Q. & Chang, C. Epigenetics in health and disease. Adv. Exp. Med. Biol. 1253, 3â55 (2020).
Helm, M. & Motorin, Y. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat. Rev. Genet. 18, 275â291 (2017).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187â1200 (2017).
Barbieri, I. & Kouzarides, T. Role of RNA modifications in cancer. Nat. Rev. Cancer 20, 303â322 (2020).
Matsumura, Y., Wei, F. Y. & Sakai, J. Epitranscriptomics in metabolic disease. Nat. Metab. 5, 370â384 (2023).
Shi, H., Wei, J. & He, C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640â650 (2019).
Ru, W. et al. Insight into m(6)A methylation from occurrence to functions. Open Biol. 10, 200091 (2020).
Lee, J. H. et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol. Cell 81, 3368â3385.e3369 (2021).
Pendleton, K. E. et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824â835.e814 (2017).
van Tran, N. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 47, 7719â7733 (2019).
Ma, H. et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 15, 88â94 (2019).
Alarcon, C. R. et al. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell 162, 1299â1308 (2015).
Alarcon, C. R., Lee, H., Goodarzi, H., Halberg, N. & Tavazoie, S. F. N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482â485 (2015).
Zhou, C. et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 20, 2262â2276 (2017).
Yang, Y. et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 27, 626â641 (2017).
Chen, Y. G. et al. N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96â109.e109 (2019).
Liu, N. et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560â564 (2015).
Yang, D. et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 46, 3906â3920 (2018).
Han, M. et al. Abnormality of m6A mRNA methylation is involved in Alzheimerâs disease. Front. Neurosci. 14, 98 (2020).
Liu, L. et al. N(6)-Methyladenosine: a potential breakthrough for human cancer. Mol. Ther. Nucleic Acids 19, 804â813 (2020).
Paramasivam, A., Vijayashree Priyadharsini, J. & Raghunandhakumar, S. N6-adenosine methylation (m6A): a promising new molecular target in hypertension and cardiovascular diseases. Hypertens. Res. 43, 153â154 (2020).
Fang, D. et al. m6A modification-mediated lncRNA TP53TG1 inhibits gastric cancer progression by regulating CIP2A stability. Cancer Sci. 113, 4135â4150 (2022).
Wang, X. et al. LncRNA FENDRR with m6A RNA methylation regulates hypoxia-induced pulmonary artery endothelial cell pyroptosis by mediating DRP1 DNA methylation. Mol. Med. 28, 126 (2022).
Xue, C., Zhao, Y. & Li, L. Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer. Biomark. Res. 8, 43 (2020).
Cui, L. et al. RNA modifications: importance in immune cell biology and related diseases. Signal Transduct. Target Ther. 7, 334 (2022).
Chen, Y. S., Yang, W. L., Zhao, Y. L. & Yang, Y. G. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA 12, e1639 (2021).
Zhang, Y. et al. 5-methylcytosine (m(5)C) RNA modification controls the innate immune response to virus infection by regulating type I interferons. Proc. Natl Acad. Sci. USA 119, e2123338119 (2022).
Yang, X. et al. RNA modifications in aging-associated cardiovascular diseases. Aging 14, 8110â8136 (2022).
Bohnsack, K. E., Hobartner, C. & Bohnsack, M. T. Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes 10, https://doi.org/10.3390/genes10020102 (2019).
Song, H. et al. Biological roles of RNA m(5)C modification and its implications in cancer immunotherapy. Biomark. Res. 10, 15 (2022).
Sun, Z. et al. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene 39, 6906â6919 (2020).
Fang, L. et al. m5C-methylated lncRNA NR_033928 promotes gastric cancer proliferation by stabilizing GLS mRNA to promote glutamine metabolism reprogramming. Cell Death Dis. 14, 520 (2023).
Elliott, B. A. et al. Modification of messenger RNA by 2â-O-methylation regulates gene expression in vivo. Nat. Commun. 10, 3401 (2019).
Erales, J. et al. Evidence for rRNA 2â-O-methylation plasticity: control of intrinsic translational capabilities of human ribosomes. Proc. Natl Acad. Sci. USA 114, 12934â12939 (2017).
van Ingen, E. et al. C/D box snoRNA SNORD113-6 guides 2â-O-methylation and protects against site-specific fragmentation of tRNA(Leu)(TAA) in vascular remodeling. Mol. Ther. Nucleic Acids 30, 162â172 (2022).
Wu, H. et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2â-O-methylation via NOP58 recruitment in colorectal cancer. Mol. Cancer 19, 95 (2020).
Mao, L. H. et al. LncRNA-LALR1 upregulates small nucleolar RNA SNORD72 to promote growth and invasion of hepatocellular carcinoma. Aging 12, 4527â4546 (2020).
Zinshteyn, B. & Nishikura, K. Adenosine-to-inosine RNA editing. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 202â209 (2009).
Gatsiou, A., Vlachogiannis, N., Lunella, F. F., Sachse, M. & Stellos, K. Adenosine-to-inosine RNA editing in health and disease. Antioxid. Redox Signal 29, 846â863 (2018).
Huang, W. et al. The snoRNA-like lncRNA LNC-SNO49AB drives leukemia by activating the RNA-editing enzyme ADAR1. Cell Discov. 8, 117 (2022).
Borchardt, E. K., Martinez, N. M. & Gilbert, W. V. Regulation and function of RNA pseudouridylation in human cells. Annu. Rev. Genet. 54, 309â336 (2020).
Lin, T. Y., Mehta, R. & Glatt, S. Pseudouridines in RNAs: switching atoms means shifting paradigms. FEBS Lett. 595, 2310â2322 (2021).
Kiss, D. J. et al. The structure-derived mechanism of box H/ACA pseudouridine synthase offers a plausible paradigm for programmable RNA editing. ACS Catal. 12, 2756â2769 (2022).
Zacchini, F. et al. Human dyskerin binds to cytoplasmic H/ACA-box-containing transcripts affecting nuclear hormone receptor dependence. Genome Biol. 23, 177 (2022).
Badis, G., Fromont-Racine, M. & Jacquier, A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA 9, 771â779 (2003).
Breznak, S. M. et al. H/ACA snRNP-dependent ribosome biogenesis regulates translation of polyglutamine proteins. Sci. Adv. 9, eade5492 (2023).
Jady, B. E., Ketele, A., Moulis, D. & Kiss, T. Guide RNA acrobatics: positioning consecutive uridines for pseudouridylation by H/ACA pseudouridylation loops with dual guide capacity. Genes Dev. 36, 70â83 (2022).
Hentze, M. W., Castello, A., Schwarzl, T. & Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 19, 327â341 (2018).
Romero-Barrios, N., Legascue, M. F., Benhamed, M., Ariel, F. & Crespi, M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 46, 2169â2184 (2018).
Galganski, L., Urbanek, M. O. & Krzyzosiak, W. J. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 45, 10350â10368 (2017).
Morais, P., Adachi, H. & Yu, Y. T. Spliceosomal snRNA epitranscriptomics. Front. Genet. 12, 652129 (2021).
Shao, C. et al. Mechanisms for U2AF to define 3â splice sites and regulate alternative splicing in the human genome. Nat. Struct. Mol. Biol. 21, 997â1005 (2014).
Tholen, J., Razew, M., Weis, F. & Galej, W. P. Structural basis of branch site recognition by the human spliceosome. Science 375, 50â57 (2022).
Agrawal, A. A. et al. An extended U2AF(65)-RNA-binding domain recognizes the 3â splice site signal. Nat. Commun. 7, 10950 (2016).
Bernard, D. et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082â3093 (2010).
Tripathi, V. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925â938 (2010).
Miao, H. et al. MALAT1 modulates alternative splicing by cooperating with the splicing factors PTBP1 and PSF. Sci. Adv. 8, eabq7289 (2022).
Bhat, P., Honson, D. & Guttman, M. Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 22, 653â670 (2021).
Fox, A. H., Nakagawa, S., Hirose, T. & Bond, C. S. Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem. Sci. 43, 124â135 (2018).
Imamura, K. et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 54, 1055 (2014).
Hirose, T. et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169â183 (2014).
Song, C. et al. Immunotherapy for Alzheimerâs disease: targeting beta-amyloid and beyond. Transl. Neurodegener. 11, 18 (2022).
Mittal, P. & Roberts, C. W. M. The SWI/SNF complex in cancerâbiology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 17, 435â448 (2020).
Reddy, D. et al. Paraspeckles interact with SWI/SNF subunit ARID1B to regulate transcription and splicing. EMBO Rep. 24, e55345 (2023).
McDonald, B. et al. Canonical BAF complex activity shapes the enhancer landscape that licenses CD8(+) T cell effector and memory fates. Immunity 56, 1303â1319.e1305 (2023).
Fujisawa, T. & Filippakopoulos, P. Functions of bromodomain-containing proteins and their roles in homeostasis and cancer. Nat. Rev. Mol. Cell Biol. 18, 246â262 (2017).
Rahnamoun, H. et al. RNAs interact with BRD4 to promote enhanced chromatin engagement and transcription activation. Nat. Struct. Mol. Biol. 25, 687â697 (2018).
Pastori, C. et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl Acad. Sci. USA 112, 8326â8331 (2015).
Pei, R. et al. JMJD6-BRD4 complex stimulates lncRNA HOTAIR transcription by binding to the promoter region of HOTAIR and induces radioresistance in liver cancer stem cells. J. Transl. Med. 21, 752 (2023).
Pistoni, M. et al. Longnoncoding RNA NEAT1 acts as a molecular switch for BRD4 transcriptional activity and mediates repression of BRD4/WDR5 target genes. Mol. Cancer Res. 19, 799â811 (2021).
Easton, A. et al. Identification and characterization of a MAPT-targeting locked nucleic acid antisense oligonucleotide therapeutic for tauopathies. Mol. Ther. Nucleic Acids 29, 625â642 (2022).
Grossi, E. et al. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat. Commun. 11, 936 (2020).
Mancini, M. et al. Involvement of transcribed lncRNA uc.291 and SWI/SNF complex in cutaneous squamous cell carcinoma. Discov. Oncol. 12, 14 (2021).
Kim, Y. K. RNA therapy: rich history, various applications and unlimited future prospects. Exp. Mol. Med. 54, 455â465 (2022).
Lim, K. H., Yang, S., Kim, S. H. & Joo, J. Y. Elevation of ACE2 as a SARS-CoV-2 entry receptor gene expression in Alzheimerâs disease. J. Infect. 81, e33âe34 (2020).
Lim, K. H., Yang, S., Kim, S. H. & Joo, J. Y. Identifying new COVID-19 receptor neuropilin-1 in severe Alzheimerâs disease patients group brain using genome-wide association study approach. Front. Genet. 12, 741175 (2021).
Feng, R., Patil, S., Zhao, X., Miao, Z. & Qian, A. RNA therapeuticsâresearch and clinical advancements. Front. Mol. Biosci. 8, 710738 (2021).
Winkle, M., El-Daly, S. M., Fabbri, M. & Calin, G. A. Noncoding RNA therapeuticsâchallenges and potential solutions. Nat. Rev. Drug Discov. 20, 629â651 (2021).
Rasul, M. F. et al. Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy. Mol. Cancer 21, 64 (2022).
Ledford, H. Is CRISPR safe? Genome editing gets its first FDA scrutiny. Nature 623, 234â235 (2023).
Antonio-Aguirre, B. & Arevalo, J. F. Treating patients with geographic atrophy: are we there yet? Int. J. Retin. Vitreous 9, 72 (2023).
Kang, C. Avacincaptad pegol: first approval. Drugs 83, 1447â1453 (2023).
Zhu, Y., Zhu, L., Wang, X. & Jin, H. RNA-based therapeutics: an overview and prospectus. Cell Death Dis. 13, 644 (2022).
Sparmann, A. & Vogel, J. RNA-based medicine: from molecular mechanisms to therapy. EMBO J. 42, e114760 (2023).
Huang, C. K., Kafert-Kasting, S. & Thum, T. Preclinical and clinical development of noncoding RNA therapeutics for cardiovascular disease. Circ. Res. 126, 663â678 (2020).
Paunovska, K., Loughrey, D. & Dahlman, J. E. Drug delivery systems for RNA therapeutics. Nat. Rev. Genet. 23, 265â280 (2022).
Zhao, Y., Shu, R. & Liu, J. The development and improvement of ribonucleic acid therapy strategies. Mol. Ther. Nucleic Acids 26, 997â1013 (2021).
Azofeifa, J. G. et al. Enhancer RNA profiling predicts transcription factor activity. Genome Res. 28, 334â344 (2018).
Marinus, T. & Incarnato, D. RNA framework for assaying the structure of RNAs by high-throughput sequencing. Methods Mol. Biol. 2284, 63â76 (2021).
Shao, D. et al. An m6A/m5C/m1A/m7G-related long non-coding RNA signature to predict prognosis and immune features of glioma. Front. Genet. 13, 903117 (2022).
Lim, K. H., Kim, S. H., Yang, S., Chun, S. & Joo, J. Y. Advances in multiplex PCR for Alzheimerâs disease diagnostics targeting CDK genes. Neurosci. Lett. 749, 135715 (2021).
Kim, S. H., Lim, K. H., Yang, S. & Joo, J. Y. Boosting of tau protein aggregation by CD40 and CD48 gene expression in Alzheimerâs disease. FASEB J. 37, e22702 (2023).
Kim, S. H. et al. Prediction of Alzheimerâs disease-specific phospholipase c gamma-1 SNV by deep learning-based approach for high-throughput screening. Proc. Natl Acad. Sci. USA 118, https://doi.org/10.1073/pnas.2011250118 (2021).
Joo, J. Y. et al. Prediction of genetic alteration of phospholipase C isozymes in brain disorders: Studies with deep learning. Adv. Biol. Regul. 82, 100833 (2021).
Yang, S., Kim, S. H., Kang, M. & Joo, J. Y. Harnessing deep learning into hidden mutations of neurological disorders for therapeutic challenges. Arch. Pharm. Res. 46, 535â549 (2023).
Lim, K. H. et al. Cryptic mutations of PLC family members in brain disorders: recent discoveries and a deep-learning-based approach. Brain 146, 1267â1280 (2023).
Kim, S.-H., Yang, S., Yang, E., Kang, M. & Joo, J.-Y. Potent of strategic approaches for tauopathies ranging from single-cell transcriptome to microbiome. Anim. Cells Syst. 27, 378â393 (2023).
Sun, P. P. et al. DeepMRMP: a new predictor for multiple types of RNA modification sites using deep learning. Math. Biosci. Eng. 16, 6231â6241 (2019).
Noviello, T. M. R., Ceccarelli, F., Ceccarelli, M. & Cerulo, L. Deep learning predicts short non-coding RNA functions from only raw sequence data. PLoS Comput. Biol. 16, e1008415 (2020).
Liang, S. et al. Rm-LR: a long-range-based deep learning model for predicting multiple types of RNA modifications. Comput. Biol. Med. 164, 107238 (2023).
Mostavi, M., Salekin, S. & Huang, Y. Deep-2â-O-Me: predicting 2â-O-methylation sites by convolutional neural networks. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2018, 2394â2397 (2018).
Zhuang, J. et al. PseUdeep: RNA pseudouridine site identification with deep learning algorithm. Front. Genet. 12, 773882 (2021).
Wang, H. et al. EMDLP: ensemble multiscale deep learning model for RNA methylation site prediction. BMC Bioinform. 23, 221 (2022).
Hasan, M. M. et al. Deepm5C: a deep-learning-based hybrid framework for identifying human RNA N5-methylcytosine sites using a stacking strategy. Mol. Ther. 30, 2856â2867 (2022).
Luo, Z. et al. DLm6Am: a deep-learning-based tool for identifying N6,2â-O-dimethyladenosine sites in RNA sequences. Int. J. Mol. Sci. 23, https://doi.org/10.3390/ijms231911026 (2022).
Wen, M., Cong, P., Zhang, Z., Lu, H. & Li, T. DeepMirTar: a deep-learning approach for predicting human miRNA targets. Bioinformatics 34, 3781â3787 (2018).
Alam, T., Islam, M. T., Househ, M., Bouzerdoum, A. & Kawsar, F. A. DeepDSSR: deep learning structure for human donor splice sites recognition. Stud. Health Technol. Inf. 262, 236â239 (2019).
Pan, X., Fang, Y., Li, X., Yang, Y. & Shen, H. B. RBPsuite: RNA-protein binding sites prediction suite based on deep learning. BMC Genom. 21, 884 (2020).
Zeng, M. et al. DeepLncLoc: a deep learning framework for long non-coding RNA subcellular localization prediction based on subsequence embedding. Brief Bioinform. 23, https://doi.org/10.1093/bib/bbab360 (2022).
Chaabane, M., Williams, R. M., Stephens, A. T. & Park, J. W. circDeep: deep learning approach for circular RNA classification from other long non-coding RNA. Bioinformatics 36, 73â80 (2020).
Chantsalnyam, T., Siraj, A., Tayara, H. & Chong, K. T. ncRDense: a novel computational approach for classification of non-coding RNA family by deep learning. Genomics 113, 3030â3038 (2021).
Fu, L. et al. UFold: fast and accurate RNA secondary structure prediction with deep learning. Nucleic Acids Res. 50, e14 (2022).
Kagaya, Y. et al. NuFold: a novel tertiary RNA structure prediction method using deep learning with flexible nucleobase center representation. bioRxiv, https://doi.org/10.1101/2023.09.20.558715 (2023).
Nasaev, S. S., Mukanov, A. R., Kuznetsov, I. I. & Veselovsky, A. V. AliNAâa deep learning program for RNA secondary structure prediction. Mol. Inform. 42, e2300113 (2023).
Niu, M., Wu, J., Zou, Q., Liu, Z. & Xu, L. rBPDL:predicting RNA-binding proteins using deep learning. IEEE J. Biomed. Health Inf. 25, 3668â3676 (2021).
Du, X., Zhao, X. & Zhang, Y. DeepBtoD: improved RNA-binding proteins prediction via integrated deep learning. J. Bioinform. Comput Biol. 20, 2250006 (2022).
Acknowledgements
J.Y.J., S.Y. and S.H.K. conceived the manuscript. S.Y. and S.H.K. created the figures and wrote the manuscript. E.Y. collected the information and edited the manuscript. M.K. revised and reviewed the manuscript. J.Y.J. supervised manuscript preparation. We thank Jeehye Jung and Jeonghyeon Choi for assisting with the data investigation. All authors have read and agreed to the published version of the manuscript. The graphical figures were made with biorender.com. This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (RS-2023-00217123) and the Ministry of Science and ICT (2022R1A2C1002984).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisherâs note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the articleâs Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the articleâs Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, S., Kim, SH., Yang, E. et al. Molecular insights into regulatory RNAs in the cellular machinery. Exp Mol Med 56, 1235â1249 (2024). https://doi.org/10.1038/s12276-024-01239-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-024-01239-6
This article is cited by
-
Regulatory RNA: from molecular insights to therapeutic frontiers
Experimental & Molecular Medicine (2024)