Rapidity of Genomic Adaptations to Prasinovirus Infection in a Marine Microalga
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture Conditions
2.2. Karyotype Analysis
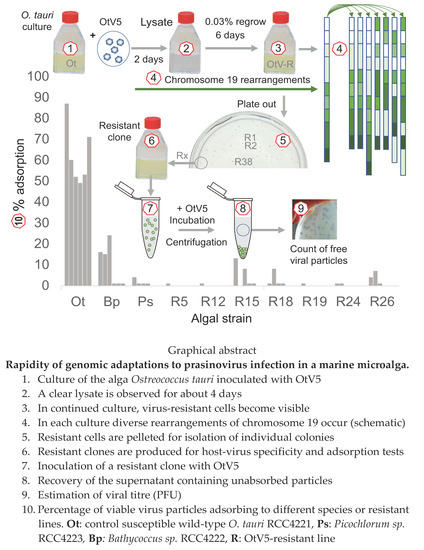
2.3. Virus Adsorption Assay
3. Results
3.1. Resistant Cells Grew 3 Days after Virus Inoculation
3.2. Evidence for Chromosomal Rearrangements within 8 Days
3.3. Acquisition of Resistance Is Frequent
3.4. OtV5-Resistant Strains Showed a Wide Spectrum of Viral Resistance
3.5. OtV5-Resistant Strains Do Not Adsorb OtV5
4. Discussion
4.1. Is Resistance Induced?
4.2. Genomic Rearrangements Occur Rapidly
4.3. Acquired Resistance Shows a Broad Specificity Spectrum
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mayer, J.A.; Taylor, F.J.R. Virus which lyses the marine nanoflagellate Micromonas pusilla. Nature 1979, 281, 299–301. [Google Scholar] [CrossRef]
- Short, S.M. The ecology of viruses that infect eukaryotic algae. Environ. Microbiol. 2012, 14, 2253–2271. [Google Scholar] [CrossRef] [PubMed]
- Hingamp, P.; Grimsley, N.; Acinas, S.G.; Clerissi, C.; Subirana, L.; Poulain, J.; Ferrera, I.; Sarmento, H.; Villar, E.; Lima-Mendez, G.; et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 2013, 7, 1678–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derelle, E.; Ferraz, C.; Escande, M.-L.; Eychenie, S.; Cooke, R.; Piganeau, G.; Desdevises, Y.; Bellec, L.; Moreau, H.; Grimsley, N. Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS ONE 2008, 3, e2250. [Google Scholar] [CrossRef] [PubMed]
- Weynberg, K.D.; Allen, M.J.; Ashelford, K.; Scanlan, D.J.; Wilson, W.H. From small hosts come big viruses: The complete genome of a second Ostreococcus tauri virus, OtV-1. Environ. Microbiol. 2009, 11, 2821–2839. [Google Scholar] [CrossRef] [PubMed]
- Weynberg, K.D.; Allen, M.J.; Gilg, I.C.; Scanlan, D.J.; Wilson, W.H. Genome sequence of Ostreococcus tauri virus OtV-2 throws light on the role of picoeukaryote niche separation in the ocean. J. Virol. 2011, 85, 4520–4529. [Google Scholar] [CrossRef] [PubMed]
- Derelle, E.; Monier, A.; Cooke, R.; Worden, A.Z.; Grimsley, N.H.; Moreau, H. Diversity of viruses Infecting the green microalga Ostreococcus lucimarinus. J. Virol. 2015, 89, 5812–5821. [Google Scholar] [CrossRef] [PubMed]
- Moreau, H.; Piganeau, G.; Desdevises, Y.; Cooke, R.; Derelle, E.; Grimsley, N. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J. Virol. 2010, 84, 12555–12563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhou, J.; Liu, T.; Yu, Y.; Pan, Y.; Yan, S.; Wang, Y. Four novel algal virus genomes discovered from Yellowstone Lake metagenomes. Sci. Rep. 2015, 5, 15131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derelle, E.; Ferraz, C.; Rombauts, S.; Rouzé, P.; Worden, A.Z.; Robbens, S.; Partensky, F.; Degroeve, S.; Echeynié, S.; Cooke, R.; et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc. Natl. Acad. Sci. USA 2006, 103, 11647–11652. [Google Scholar] [CrossRef] [PubMed]
- Blanc-Mathieu, R.; Verhelst, B.; Derelle, E.; Rombauts, S.; Bouget, F.-Y.; Carré, I.; Château, A.; Eyre-Walker, A.; Grimsley, N.; Moreau, H.; et al. An improved genome of the model marine alga Ostreococcus tauri unfolds by assessing Illumina de novo assemblies. BMC Genom. 2014, 15, 1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worden, A.; Lee, J.-H.; Mock, T.; Rouzé, P.; Simmons, M.P.; Aerts, A.L.; Allen, A.E.; Cuvelier, M.L.; Derelle, E.; Everett, M.V.; et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Palenik, B.; Grimwood, J.; Aerts, A.; Rouze, P.; Salamov, A.; Putnam, N.; Dupont, C.; Jorgensen, R.; Derelle, E.; Rombauts, S.; et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 2007, 104, 7705–7710. [Google Scholar] [CrossRef] [PubMed]
- Moreau, H.; Verhelst, B.; Couloux, A.; Derelle, E.; Rombauts, S.; Grimsley, N.; Bel, M.V.; Poulain, J.; Katinka, M.; Hohmann-Marriott, M.F.; et al. Gene functionalities and genome structure in Bathycoccus prasinos reflect cellular specializations at the base of the green lineage. Genome Biol. 2012, 13, R74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derelle, E.; Yau, S.; Moreau, H.; Grimsley, N.H. Prasinovirus Attack of Ostreococcus Is Furtive by Day but Savage by Night. J. Virol. 2018, 92, e01703-17. [Google Scholar] [CrossRef] [PubMed]
- Monier, A.; Chambouvet, A.; Milner, D.S.; Attah, V.; Terrado, R.; Lovejoy, C.; Moreau, H.; Santoro, A.E.; Derelle, É.; Richards, T.A. Host-derived viral transporter protein for nitrogen uptake in infected marine phytoplankton. Proc. Natl. Acad. Sci. USA 2017, 114, E7489–E7498. [Google Scholar] [CrossRef] [PubMed]
- Monier, A.; Welsh, R.M.; Gentemann, C.; Weinstock, G.; Sodergren, E.; Armbrust, E.V.; Eisen, J.A.; Worden, A.Z. Phosphate transporters in marine phytoplankton and their viruses: cross-domain commonalities in viral-host gene exchanges. Environ. Microbiol. 2012, 14, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Grimsley, N.; Escande, M.; Subirana, L.; Derelle, E.; Moreau, H. Acquisition and maintenance of resistance to viruses in eukaryotic phytoplankton populations. Environ. Microbiol. 2011, 13, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Yau, S.; Hemon, C.; Derelle, E.; Moreau, H.; Piganeau, G.; Grimsley, N. A Viral Immunity Chromosome in the Marine Picoeukaryote, Ostreococcus tauri. PLoS Pathog. 2016, 12, e1005965. [Google Scholar] [CrossRef] [PubMed]
- Courties, C.; Vaquer, A.; Troussellier, M.; Lautier, J.; Chretiennotdinet, M.J.; Neveux, J.; Machado, C.; Claustre, H. Smallest Eukaryotic Organism. Nature 1994, 370, 255. [Google Scholar] [CrossRef]
- Courties, C.; Perasso, R.; Chretiennot-Dinet, M.J.; Gouy, M.; Guillou, L.; Troussellier, M. Phylogenetic analysis and genome size of Ostreococcus tauri (Chlorophyta, Prasinophyceae). J. Phycol. 1998, 34, 844–849. [Google Scholar] [CrossRef]
- Clerissi, C.; Desdevises, Y.; Grimsley, N.H. Prasinoviruses of the marine green alga Ostreococcus tauri are mainly species-specific. J. Virol. 2012, 86, 4611–4619. [Google Scholar] [CrossRef] [PubMed]
- Trask, B.J.; van den Engh, G.J.; Elgershuizen, J.H. Analysis of phytoplankton by flow cytometry. Cytometry 1982, 2, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, F.; Derelle, E.; Guillou, L.; Le Gall, F.; Vaulot, D.; Moreau, H. Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae). Environ. Microbiol. 2005, 7, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Meints, R.H.; Burbank, D.E.; Van Etten, J.L.; Lamport, D.T. Properties of the Chlorella receptor for the virus PBCV-1. Virology 1988, 164, 15–21. [Google Scholar] [CrossRef]
- Bellec, L.; Clerissi, C.; Edern, R.; Foulon, E.; Simon, N.; Grimsley, N.; Desdevises, Y. Cophylogenetic interactions between marine viruses and eukaryotic picophytoplankton. BMC Evol. Biol. 2014, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Krasovec, M.; Vancaester, E.; Rombauts, S.; Bucchini, F.; Yau, S.; Hemon, C.; Lebredonchel, H.; Grimsley, N.; Moreau, H.; Sanchez-Brosseau, S.; et al. Genome analyses of the microalga Picochlorum provide insights into the evolution of thermotolerance in the green lineage. Genome Biol. Evol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Krasovec, M.; Eyre-Walker, A.; Sanchez-Ferandin, S.; Piganeau, G. Spontaneous Mutation Rate in the Smallest Photosynthetic Eukaryotes. Mol. Biol. Evol. 2017, 34, 1770–1779. [Google Scholar] [CrossRef] [PubMed]
- Subirana, L.; Péquin, B.; Michely, S.; Escande, M.-L.; Meilland, J.; Derelle, E.; Marin, B.; Piganeau, G.; Desdevises, Y.; Moreau, H.; et al. Morphology, Genome Plasticity, and Phylogeny in the Genus Ostreococcus Reveal a Cryptic Species, O. mediterraneus sp. nov. (Mamiellales, Mamiellophyceae). Protist 2013, 164, 643–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanc-Mathieu, R.; Krasovec, M.; Hebrard, M.; Yau, S.; Desgranges, E.; Martin, J.; Schackwitz, W.; Kuo, A.; Salin, G.; Donnadieu, C.; et al. Population genomics of picophytoplankton unveils novel chromosome hypervariability. Sci. Adv. 2017, 3, e1700239. [Google Scholar] [CrossRef] [PubMed]
- Bellec, L.; Grimsley, N.; Derelle, E.; Moreau, H.; Desdevises, Y. Abundance, spatial distribution and genetic diversity of Ostreococcus tauri viruses in two different environments. Environ. Microbiol. Rep. 2010, 2, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Heath, S.E.; Knox, K.; Vale, P.F.; Collins, S. Virus Resistance Is Not Costly in a Marine Alga Evolving under Multiple Environmental Stressors. Viruses 2017, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Vannier, T.; Leconte, J.; Seeleuthner, Y.; Mondy, S.; Pelletier, E.; Aury, J.-M.; de Vargas, C.; Sieracki, M.; Iudicone, D.; Vaulot, D.; et al. Survey of the green picoalga Bathycoccus genomes in the global ocean. Sci. Rep. 2016, 6, 37900. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yau, S.; Caravello, G.; Fonvieille, N.; Desgranges, É.; Moreau, H.; Grimsley, N. Rapidity of Genomic Adaptations to Prasinovirus Infection in a Marine Microalga. Viruses 2018, 10, 441. https://doi.org/10.3390/v10080441
Yau S, Caravello G, Fonvieille N, Desgranges É, Moreau H, Grimsley N. Rapidity of Genomic Adaptations to Prasinovirus Infection in a Marine Microalga. Viruses. 2018; 10(8):441. https://doi.org/10.3390/v10080441
Chicago/Turabian StyleYau, Sheree, Gaëtan Caravello, Nadège Fonvieille, Élodie Desgranges, Hervé Moreau, and Nigel Grimsley. 2018. "Rapidity of Genomic Adaptations to Prasinovirus Infection in a Marine Microalga" Viruses 10, no. 8: 441. https://doi.org/10.3390/v10080441