Abstract
The Human Connectome Project (HCP) is an ambitious 5-year effort to characterize brain connectivity and function and their variability in healthy adults. This review summarizes the data acquisition plans being implemented by a consortium of HCP investigators who will study a population of 1200 subjects (twins and their non-twin siblings) using multiple imaging modalities along with extensive behavioral and genetic data. The imaging modalities will include diffusion imaging (dMRI), resting-state fMRI (R-fMRI), task-evoked fMRI (T-fMRI), T1- and T2-weighted MRI for structural and myelin mapping, plus combined magnetoencephalography and electroencephalography (MEG/EEG). Given the importance of obtaining the best possible data quality, we discuss the efforts underway during the first two years of the grant (Phase I) to refine and optimize many aspects of HCP data acquisition, including a new 7T scanner, a customized 3T scanner, and improved MR pulse sequences.
Keywords: Connectivity, fMRI, Diffusion imaging, MEG/EEG, Twins, Behavior
Introduction
Recent advances in neuroimaging, including many that are discussed in this special issue, have made it feasible to examine human brain connectivity systematically and across the whole brain in large numbers of individual subjects. Progress in the nascent field of connectomics led NIH in 2009 to announce a Request for Applications for the Human Connectome Project (HCP), with an overarching objective of studying human brain connectivity and its variability in healthy adults. In September, 2010, grants were awarded to two consortia (http://www.neuroscienceblueprint.nih.gov/connectome/). One is a 5-year grant to a consortium of ten institutions in the United States and Europe, led by Washington University and the University of Minnesota (the ‘WU-Minn HCP Consortium’). This consortium aims to study brain connectivity and function with a genetically-informative design in 1200 individuals using four MR-based modalities plus MEG and EEG. Behavioral and genetic data will also be acquired from these subjects. The second is a 3-year grant to a consortium led by Harvard/MGH and UCLA to develop an advanced MR scanner for diffusion imaging.
A deeper understanding of human brain connectivity and its variability will provide valuable insights into what makes us uniquely human and what accounts for the great diversity of behavioral capacities and repertoires in healthy adults. It will provide a critical baseline of knowledge for future studies of brain connectivity during development and aging and in myriad neurodevelopmental, neuropsychiatric and neurological disorders. Also, the data acquisition strategies and analysis methods developed under the auspices of the HCP will be freely shared and will benefit many other projects. Increasing both the commonality and the sensitivity of methods used to characterize human brain connectivity across different studies will enhance our ability to detect subtle links between genetics, human brain connectivity patterns, and behavioral variation.
Despite their great promise, all of the modalities that can be applied to in vivo human connectomics currently have serious limitations in their sensitivity, accuracy, and resolution (Van Essen and Ugurbil, 2012). Hence, during Phase I of the grant (until the summer of 2012) the WU-Minn HCP consortium is making a major effort to improve the methods of data acquisition and analysis. This includes a new 3T MRI scanner designed to improve the quality and resolution of connectivity data, as well as a new 7T scanner, both of which will capitalize on major improvement in MR pulse sequences. This initial phase will be followed by a 3-year period of data acquisition from the main cohort (Phase II). The combination of methods refinement followed by extensive data acquisition makes the HCP a unique enterprise compared to several other large-scale imaging efforts that are also underway (see Discussion).
This review focuses on the data acquisition aspects of the HCP, given their critical importance for the endeavor. After a brief overview of the HCP objectives, we describe the subject cohort and behavioral measures, followed by the hardware configuration and data acquisition strategies for each of the main imaging modalities. Already there have been significant methodological advances that provide grounds for optimism about the data quality that will be attainable. Approaching near-optimal solutions will be very challenging given the large number of factors and parameters needing evaluation. We provide examples of our general approach to this problem.
Overview of the HCP
Fig. 1 provides a high-level view of our plans for data acquisition in Phase II of the project. Data will be acquired from 1200 subjects, comprising young adult sibships of average size 3–4, including twins and their non-twin siblings. Each subject will spend 2 days at WashU for behavioral assessment, blood draw for eventual genotyping, and multiple MR scanning sessions (4 sessions, with 3 lasting 1 h). The WashU scans will be carried out using a customized 3T Connectome Scanner adapted from a Siemens Skyra (Siemens AG, Erlanger, Germany); a subset of 200 subjects will also be scanned at UMinn using a new 7T scanner (MR hardware section). On both the 3T and 7T systems, the MR scans will use advanced pulse sequences to acquire dMRI, R-fMRI, and T-fMRI, plus T1w and T2w anatomical scans. T-fMRI scans will include a range of tasks aimed at providing broad coverage of the brain and identifying as many functionally distinct domains and cortical parcels as possible.
Fig. 1.
Schematic summary for acquiring imaging, behavioral, and genetic data using MR and MEG/EEG scanners at three HCP data acquisition sites. Left: Behavioral testing, blood draws for genotyping, and scanning on a 3T Skyra will be carried out on 1200 healthy adults at Washington University (WashU). Center: Major data acquisition modalities are indicated in the center column; for task-fMRI and behavior, major domains are listed. Top right: A subset of 200 subjects will be scanned on a 7T Skyra at the University of Minnesota (UMinn). Bottom right: A subset of 100 subjects will be scanned using magnetoencephalography (MEG) and perhaps electroencephalography (EEG) at St. Louis University (SLU).
A subset of 100 subjects will also be studied with combined MEG/EEG at St. Louis University (SLU); if possible, some of these will be in the group also scanned at 7T. MEG and EEG provide much better temporal resolution (milliseconds instead of seconds) but lower spatial resolution than MR (MEG/EEG section).
The behavioral measures will span a broad range in the domains of cognition, emotion, perception, and motor function (Behavioral measures section). They will be drawn mainly from the NIH Toolbox but will be supplemented by a number of complementary additional measures. Blood samples from all subjects will be used for genotyping in year 5, at which time full-genome sequencing may be affordable (Genetics section).
Extensive efforts to refine many aspects of data analysis are underway for each modality, as will be discussed in future publications. Another major thrust is to implement a robust and user-friendly informatics platform to support data management and data mining (Marcus et al., 2011).
In principle, it would be valuable to collect data from additional noninvasive0020imaging modalities (e.g., PET and NIRS). However, given overall budget constraints this would require reducing the total number of subjects studied. The strategy we adopted reflects a trade-off and balance between (i) acquiring as much information as is feasible using multiple modalities related to brain connectivity and function, and (ii) having a subject population sufficiently large to systematically explore the neurobiological and genetic bases of individual variability in brain circuitry and behavioral phenotype.
Study subjects
A key objective is to understand inter-individual variability of brain circuits, including its genetic bases and its relation to behavior, rather than merely aiming to determine the average, or typical connectivity in healthy adults. This will be achieved by sampling 300–400 young adult sibships of average size 3–4, with most of these sibships including a MZ or DZ twin pair. All subjects will be between 22 and 35 years old, an age range chosen to represent healthy adults beyond the age of major neurodevelopmental changes and before the onset of neurodegenerative changes. While the HCP will be cross-sectional, many participants will be drawn from ongoing longitudinal studies (Sartor et al., 2011; Edens et al., 2010); they will have extensive previous assessments, particularly with respect to history of the presence or absence of emotional and behavioral problems. This will allow us to recruit a sample of relatively healthy individuals free of a prior history of significant psychiatric or neurological illnesses. Our goal is to capture a broad range of variability in healthy individuals with respect to behavioral, ethnic, and socioeconomic diversity. We will define ‘healthy’ broadly, to avoid having an unduly narrow ‘supernormal’ case series that might not be representative of the population at large. We will exclude sibships with individuals having severe neurodevelopmental disorders (e.g. autism), documented neuropsychiatric disorders (e.g. schizophrenia or severe recurrent depression) or neurologic disorders (e.g. Parkinson's disease), but will include individuals who are smokers, are overweight, or have a history of heavy drinking or recreational drug use without having experienced severe symptoms (Supplemental Table S1 lists the full set of inclusion and exclusion criteria under consideration). This strategy will enable future connectivity studies on psychiatric patients, many of whom smoke, are overweight, or have subclinical substance use behaviors, to be compared to connectivity data on HCP ‘healthy individuals’ having similar profiles. Twins born prior to 34 weeks gestation and non-twins born prior to 37 weeks gestation will be excluded. This acknowledges the higher incidence of prematurity in twins and focuses on exclusion of individuals born very prematurely. Our initial screening will include a detailed questionnaire developed explicitly for the HCP to determine presence or absence of the inclusion/exclusion criteria. This will be followed by an additional extensive, reliable, and valid psychiatric interview, the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA, Bucholz et al., 1994), to confirm the absence of significant psychiatric illness. This will also allow us to include information about subthreshold psychiatric symptoms in the database, as analyses of such data may be of interest to many researchers.
The utility of twin pairs in furthering our understanding of the causes of human variation extends beyond estimating the contribution of genetic differences to individual variation (for classic early studies, see Eaves, 1982 and Martin et al., 1997; for a discussion of statistical analysis approaches, see Neale and Cardon, 1992). MZ twinning occurs randomly, so MZ twin pairs should capture the full range of genetic variability in a population. These twin pairs are genetically nearly identical; while they may share many aspects of rearing history and socioeconomic background, they also have within-pair variance due to differences in environmental exposures, stochastic processes and measurement error. Accordingly, assessment of MZ twin pairs on its own is valuable in three distinct respects. (i) It provides a within-pair contrast for effects of environmental exposure or physical or physiologic state (e.g. in pairs discordant for smoking, overweight/obesity, or diabetes). (ii) It provides a lower-bound estimate of the test–retest reliability of various HCP measures. (It is a lower bound because it reflects only genetic effects plus environmental effects shared by the twin pairs; however, it is especially valuable in experiments that for technical reasons are non-repeatable.) (iii) It provides an estimate of the covariance structure of multiple measures that is uncontaminated by individual-specific stochastic and measurement error effects.
Dizygotic twin pairs are as genetically related as ordinary full siblings, but they share their childhood environment to a much greater extent than do siblings of different ages. When added to MZ twin data, DZ twin data thus allow estimation of the extent to which genotype, shared environment, and non-shared influences each contribute to variation in traits. In multivariate analysis, this extends to understanding why traits A and B co-vary. The inclusion of additional siblings along with twins provides a further increase in statistical power for resolving genetic and environmental influences (Posthuma and Boomsma, 2000). These basic applications may be elaborated to test for genotype × environment interaction effects, where genetic influences are modified as a function of environmental exposure or experimental manipulation; conditional effects (e.g. how smoking status may affect connectivity patterns); and to test for certain strong directional models (event A leads to event B, rather than vice versa) (Neale and Cardon, 1992).
Genetics
Participants will provide blood samples that will be used to create cell-lines and for DNA extraction, with these resources available to other qualified investigators. In the final year of the project, we will genotype samples from all study participants. The genotyping method will be chosen from those available at that time, with the goal of obtaining the maximum amount of data given budgetary constraints; this may include full-genome sequencing. HCP genetic data will allow investigators to look for the effects of specific genetic variants (as identified in powerful large-scale genome-wide association studies of clinical or behavioral phenotypes) on brain connectivity patterns in healthy adults. As one example, it will be interesting to see whether differences in brain connectivity patterns are associated with genetic variants that contribute to the risk of developing Alzheimer's disease later in life (e.g. ApoE e4). The HCP data may also enable direct discovery of gene variants that affect brain connectivity patterns, especially if the HCP core protocol is replicated across multiple studies worldwide. Overall, our use of a twin-family study paradigm to analyze individual variation in brain connectivity will facilitate progress in understanding the genetic bases of individual differences in connectivity, and their covariation with normal behavior.
Behavioral measures
HCP's behavioral measures will provide important phenotypic data to compare with brain imaging and genetics. Our goal is to cover as many domains of behavior as feasible within 2–3 h of testing outside of the scanner. Our base set of assessment tools will be the NIH Toolbox, which is being developed as a brief, well-validated assessment of the domains of cognition, emotion, motor function and sensation that can be used with healthy individuals from childhood through older age (see http://www.nihtoolbox.org). This will include domains of cognition (verbal IQ, working memory, executive function, attention, language, and processing speed), emotion (negative affect, positive affect, stress and coping, and social relationships), motor function (locomotion, dexterity, strength, and endurance), and sensation (hearing, taste, touch and smell). To facilitate cross-project comparisons, we plan to incorporate additional measures similar or identical to those used by other large-scale data acquisition projects measuring brain function, structure, and connectivity that are non-overlapping with the NIH-Toolbox measures. These include measures of attention, episodic memory, visual spatial processing, and emotional face processing as used by Gur et al. (2010); the Achenbach Adult Self Report (Achenbach et al., 2005), as used in the NKI-Rockland project (http://fcon_1000.projects.nitrc.org/indi/pro/nki.html); and a variant of matrix reasoning as a measure of fluid intelligence and the NEO-FFI-60 measure of personality (McCrae and Costa, 2004), as used in a study on cognitive aging (R. Buckner, personal communication). Finally, we plan to include the Farnsworth test of color vision, the Mars test of visual contrast sensitivity, the EVA test of visual acuity, and a measure of impulsivity (delay discounting) (Estle et al., 2006). Supplemental Table S2 lists all measures we plan to acquire (Toolbox and non-Toolbox).
The broad spectrum of behavioral information acquired from all HCP subjects will enable many types of comparison and correlation between behavior and brain connectivity (functional, structural, and electrophysiological). For example, behavioral measures can be used to identify factors or eigenvectors of common variability across subjects, which are then correlated with measures of connectivity. This can be done within a cognitive domain, as in working memory (e.g. Hampson et al., 2006), or across domains and connectivity patterns, as in comparing motor behavior to measures of connectivity across networks such as motor and attention (Carter et al., 2010). An alternative strategy is to test whether specific patterns of brain connectivity co-vary in a meaningful way with behavioral measures. For example, some studies have emphasized a correlation with global measures of connectivity (Chiang et al., 2009; van den Heuvel et al., 2009). It will be important to explore how behavioral performance relates to a variety of connectivity measures, including: ‘dense connectome’ representations at the level of voxels and surface vertices; ‘parcellated connectome’ representations of connectivity between cortical and subcortical parcels defined anatomically and/or functionally; different approaches for estimating the connectivities themselves (e.g., “functional” vs. “effective” connectivity measures (Friston et al., 2003)); and graph-theoretical representations at the level of brain networks and subnetworks (Bullmore and Sporns, 2009). Accordingly, it is important that the HCP informatics platform provides access to connectivity data at each major level of analysis, including voxelwise time-course data (Marcus et al., 2011).
MR hardware
To obtain the best possible MR data quality while scanning many subjects for the HCP, we decided to pursue a dual path involving customized 3T and 7T scanners. 3T systems are the more mature and robust platforms, compatible with the need to scan a large number of subjects. 7T systems offer advantages, especially for the resting and task-based fMRI studies, but also for diffusion-based techniques if sufficiently short echo times can be achieved for diffusion weighting. However, 7T platforms are less mature and more challenging to work with, and are thus incompatible with an ambitious data collection strategy. Accordingly, our plan is to scan all 1200 subjects at 3T, and 200 of them also at 7T. Both scanners will be modified to improve performance compared to what is available on a standard platform. There is also a possibility of imaging some HCP subjects using a new 10.5T whole body scanner that the CMRR at UMinn is building through support from a separate NIH grant. However, whether the HCP is able to scan at this ultrahigh field will depend on when the system becomes operational and key scanning protocols implemented.
New Connectome 3T scanner
Our design for the Connectome 3T MRI scanner took into consideration issues of reliability, subject comfort, and potential risks inherent in new hardware development. Unique features of the Connectome 3T involve the gradients and the RF-receive hardware. Diffusion imaging (dMRI) benefits from high gradient amplitudes that can shorten the diffusion encoding period and thus increase SNR. Multichannel receive capability is critical to parallel imaging techniques that are being developed in this project to significantly reduce whole brain data acquisition times both for fMRI and for dMRI (see below).
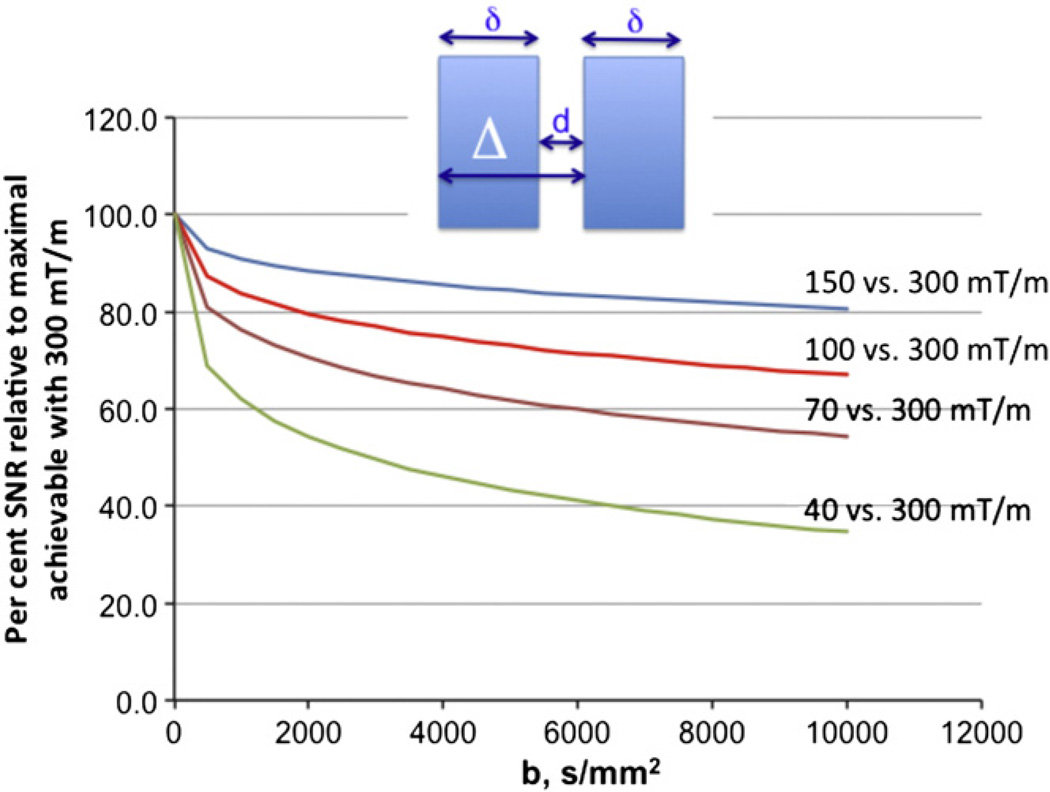
We considered several options for achieving gradient amplitudes higher than the 40 mT/m available on standard Siemens 3T scanners. Over the range from 40 mT/m to 300 mT/m the SNR gains depend nonlinearly on the b-value as well as the gradient strength. Fig. 2 demonstrates simulated SNR values achievable with a Stejskal–Tanner pulsed gradient diffusion sequence modeled assuming infinite slew gradients. Due to the sequence's G2 non-linear dependence of b-value, stronger gradients (G) do not proportionately reduce pulse width (Tp) or the minimum possible echo time (TE) on which SNR is dependent.1 The relative SNR (normalized to 100% for 300 mT/m) depends on the b-value. However, even for very ambitious b values (104 s/mm2), 100 mT/m maximum gradient strength provides ~70% of the SNR achievable relative to a 300 mT/m maximum.
Fig. 2.
Relative SNR at the central k space point in diffusion imaging with 150, 100, 70, and 40 mT/m maximum gradients relative to maximum achievable with 300 mT/m when TE is minimized using the available gradient amplitude, calculated for white matter at different b-values ranging from 500 to 10,000.
Based on these considerations, we chose a gradient configuration that can achieve a maximum gradient strength of 100 mT/m using existing and tested hardware components. Specifically, we are using a Siemens 3T Skyra scanner modified to include a Siemens SC72 gradient coil that has been used extensively in 7T scanners, where its maximum gradient strength is 70 mT/m. This will be further increased to ~ 100 mT/m using gradient amplifiers with higher current output, adapted from the Siemens 1.5T Aera scanner. This design entails only low technical risk and is well suited to our HCP objectives.
Alternative available de novo designs that theoretically could approach 300 mT/m are technically demanding and at risk of not meeting key performance characteristics (e.g. eddy currents, nonlinearities, stability, duty cycle, safety etc.). The SC72 has excellent eddy current performance in its standard configuration in an 82 cm bore magnet and should perform even better in the 90 cm bore 3T magnet. The Skyra scanner has 64 receiver channels, for use with a commercial 32-channel head coil and with customized arrays having larger number of coils that will be designed at CMRR and explored for improved SNR and acceleration.
7T scanner
The (new) UMinn 7T is also equipped with SC72 gradients and will have 32 channels initially, but will be upgraded to 64 channels before 7T scanning on the main cohort commences. The system will have third-order shims, which will improve EPI quality. RF coils will consist of multichannel receive and transmit arrays to be built at CMRR.
MR data acquisition
Important advances in pulse sequences will benefit three MR modalities (dMRI, R-fMRI, and T-fMRI) and are described in Pulse sequence improvements section. This description is followed by subsections on modality-specific aspects of MR data acquisition.
Pulse sequence improvements
The primary approach to fMRI and diffusion imaging for connectivity studies involves single shot imaging using EPI. Since its initial application, EPI scan times for whole brain coverage have not substantially decreased. Progress in shortening the EPI acquisition time for spatial encoding (Pruessmann et al., 1999; Sodickson et al., 1999; Griswold et al., 2002; Liang et al., 2003) only modestly reduces acquisition time for whole brain coverage. This modest reduction is because each slice incorporates a physiological contrast preparation period that can equal or exceed the time employed for collecting the EPI echo train. A major objective of the HCP is to achieve rapid whole-brain image acquisition with high spatial resolution for both diffusion imaging and fMRI.
Our approach to reducing scan time capitalizes on the simultaneous excitation of multiple brain slices and sharing diffusion or BOLD preparation among all slices excited. This is accomplished with multiple receivers and multiband excitations (Larkman et al., 2001), as developed for fMRI by the UMinn group (Moeller et al., 2010), and with SIR, involving acquisitions of multiple slices adjacent in time but in the same echo train (Feinberg et al., 2002). These can be combined into Multiplexed EPI (Feinberg et al., 2010). Acquiring many slices in the time of a single EPI echo train (or marginally longer echo train when SIR is employed) and a single contrast preparation period, permits sub-second whole brain coverage at 2 or 3 mm isotropic resolution (Fig. 3), yielding improved resting state fMRI results (see R-fMRI acquisition strategies section), and substantially reduced acquisition times for dMRI. These advances will benefit both diffusion and fMRI data directly through higher data acquisition rates, without serious losses in SNR, and indirectly, by reducing the total number of diffusion gradient pulses per whole brain scan, allowing more time for gradient coil cooling when very high b-values are used.
Fig. 3.
The M-EPI pulse sequence compared with conventional EPI. Top left: EPI pulse sequence generates a single slice image during each readout train, which is repeated for each slice to scan the whole brain. The multiband technique replaces the single slice excitation pulse with multiband (MB) pulses to excite several slices simultaneously, which are then unaliased using array coil sensitivity profiles. As such, far fewer repeats are required to scan the whole brain. Bottom left: Multiplexed-EPI (M-EPI) pulse sequence combines the SIR approach with the MB technique: SIR consecutively excites s slices (s = 3 is shown in the pulse sequence diagram with pulses in red, blue and green) and reads them out in a single echo train, separated in time. Using MB pulses to simultaneously excite m slices instead of exciting each single slice in the SIR approach produces the M-EPI sequence, with a “slice acceleration” of (s × m) leading to (s × m) number of slices collected in a single echo train. Right: Each column shows four (of 60) slices from a whole brain (2 mm isotropic resolution) 3T data set obtained with the M-EPI technique, shown with the (s × m) acceleration factors ranging from 4 to 12.
Adapted with permission from Feinberg et al. (2010).
Another important technical consideration involves various distortions that can plague subsequent analyses if not adequately corrected. Field map scans will be acquired and used to correct fMRI images for distortions arising from magnetic field inhomogeneities. For dMRI, pulse sequences that traverse k space in opposite phase encoding directions will be acquired and used to calculate and eliminate the image distortions (Andersson et al., 2003).
dMRI strategies
The MR hardware and pulse sequence developments described above have significant implications for the diffusion imaging strategies to be used by the HCP. Accelerated imaging will enable collection of many hundreds or even thousands of diffusion-encoded data points per voxel. The customized gradient coils on the Connectome 3T will enable acquisition of high b-value data while reducing the usual SNR trade-off. Because such data have not previously been acquired in human subjects, Phase I of the HCP will entail extensive piloting and testing by the diffusion imaging team on both 3T and 7T datasets.
We aim to identify a diffusion imaging acquisition and reconstruction protocol that will (a) provide veridical reconstructions of fiber orientations in a physical phantom; (b) provide high multi-orientation sensitivity and low uncertainty in regions of crossing fibers in vivo; (c) provide high test–retest reliability over the whole brain; and (d) provide accurate connectivity data when compared to expectations from macaque tracer studies and from same-subject functional connectivity derived from R-fMRI (R-fMRI acquisition strategies section). Among the many decisions that must be made, the most significant are the choice of diffusion-encoding scheme, for maximizing orientation sensitivity, and the choice of spatial resolution, which involves a trade-off between the accuracy of orientation peaks and the sensitivity to crossing fibers and minor pathways. We will evaluate and compare diffusion encoding schemes that sample k-space using single or multiple spherical shells, with the parameters of each scheme pre-optimized. Testing on the customized 3T Skyra, which commenced in the fall of 2011, will aim to efficiently narrow down the primary choices using multiple criteria as described above. This will be followed by fine-tuning of acquisition parameters.
In conjunction with data acquisition improvements, we are performing extensive evaluation and optimization of diffusion imaging reconstruction methods. The availability of high resolution and high SNR data will open up new possibilities. For example, we are extending multi-fiber fitting algorithms to account for (i) more complex fiber architectures, such as fanning and bending fibers and (ii) more complex data types, such as multi-q-shell or Cartesian acquisitions (Aganj et al., 2010). These new techniques will be evaluated against established techniques such as compartment modeling (Behrens et al., 2007), spherical deconvolution (Tournier et al., 2004), and Diffusion Spectrum Imaging reconstructions (Wedeen et al., 2008).
R-fMRI acquisition strategies
As illustrated already (Pulse sequence improvements section), important advances in pulse sequences have emerged from early HCP efforts. This includes combining two EPI accelerations that in combination markedly reduce TR (Feinberg et al., 2010). The reduction in TR (to less than a half second, i.e., much less than T1) decreases the SNR in each individual fMRI image, but with respect to final time series statistics, the increased number of timepoints more than compensates for this. The expected overall SNR change is a gain of 10–15%. However, for high-dimensional multiple regressions (such as that implicit in a high-dimensional functional parcellation using independent component analysis), we found an increase in effective SNR of 60% when reducing TR from 2.5 s to 0.4 s, because of the importance of the temporal degrees of freedom in such analysis. A similar gain (and for similar reasons) may occur in some network modeling analyses, such as those involving partial correlation (Smith et al., 2011) to estimate ‘direct’ network connections.
Additional increases in acceleration factors are anticipated, but they are likely to yield diminishing returns, because distortions and reconstruction artifacts may increase, while the temporal sampling becomes much faster than useful temporal information available in the (hemodynamically blurred) fMRI timeseries. On the other hand, there may be additional valuable gains, including an improved ability to model and remove physiological artifacts (Glover et al., 2000) including head motion (Power et al., 2012); improved ability to model nonstationarities (temporal variation) in the network structure (Chang and Glover, 2010); improvement in estimating higher-order statistics for network modeling (Shimizu et al., 2006); and richer modeling of the temporal dynamics of R-fMRI fluctuations and in the interactions between different functional areas (Smith et al., 2012).
As with dMRI, the effort to optimize R-fMRI acquisition parameters for Phase II data acquisition will require choices among many competing factors that will differ for 3T and 7T scanners. It will entail careful choice of pulse sequence parameters along with ‘standard’ parameters such as spatial and temporal resolution, echo-time (TE), bandwidth, MB and SIR slice acceleration factors, and within-slice parallel acceleration factor (which have different effects on g-factors and the use of partial-k-space). The interdependencies can be complex, and the choices for single parameters can involve tradeoffs. For example, one TE might give better overall SNR, whereas a different value might show better signal localization in tissue vs. local larger veins. A key objective will be to achieve sub-second TR while minimizing EPI distortion and dropout, and maximizing SNR and spatial resolution. Endpoints by which the results will be judged will include maximization of the number of functional parcels that can be reproducibly distinguished from one another, as well as the reproducibility of the network connections (between these parcels) that are then estimated. These R-fMRI distinctions can also be related to functional distinctions (Smith et al., 2009). Other decisions involve different kinds of tradeoffs: for example, the longer the imaging session the better, from the point of view of imaging data quality and the ability to sample dynamics of functional connectivity. However, this must be balanced against subjects' compliance and load, given the many modalities of data acquisition.
T-fMRI acquisition strategies
Our primary goals in including task-related fMRI measures (T-fMRI) are to (i) help identify as many “nodes” (functionally distinct brain parcels) as possible that can guide, validate, and interpret the results of the connectivity analyses that will be conducted on R-fMRI and dMRI data; (ii) provide task-activation data that can be combined with MEG data to better understand information flow within networks; (iii) allow comparison of network connectivity in a task context to connectivity results generated using R-fMRI; and (iv) to understand the relative utility of T-fMRI and R-fMRI in predicting individual differences in behavior and genetic influences. To accomplish these goals, we are developing a battery of tasks that can identify node locations in as wide a range of neural systems as feasible within realistic time constraints (~60 min in Phase II). In Phase I, we are piloting a larger number of tasks than we anticipate being able to use in Phase II. We will compare the sensitivity, reliability and brain coverage afforded by these tasks to arrive at a final T-fMRI battery that balances optimizing the psychometric properties of the activation measures (i.e., high reliability and sensitivity are necessary for individual difference and genetic analyses) with behavioral validity and interpretability. Phase I piloting includes measures of visual-motor processes (retinotopy, motor strip mapping, biological and non-biological motion), as well as a range of cognitive (working memory, episodic memory, language, attention, stimulus category representations) and affective/social processes (emotion recognition, reward and punishment based decision making, and social cognition). When possible, we are piloting tasks that allow us to assess multiple networks simultaneously. For example, we have developed a working memory task that uses different categories of stimuli. This enables collapsing across stimulus type to identify working memory related networks, and separately collapsing across memory loads to identify brain regions that respond differentially to different stimulus types. In choosing tasks to pilot in Phase 1, we emphasized ones with existing evidence of suitability as localizers in individual subjects, or evidence for their reliability across subjects or within subjects across time. We also emphasized paradigms suitable for optimized blocked designs to achieve maximum efficiency. Supplemental Table S3 lists the tasks currently being piloted.
Like R-fMRI, T-fMRI is likely to benefit considerably from low-TR data acquisition. For example, improved temporal resolution should aid in discerning differences in the time course of task activation/deactivation according to brain region and/or task (e.g., Nelson et al., 2010). The choice of T-fMRI pulse sequence parameters along with ‘standard’ parameters such as spatial and temporal resolution will involve many of the same considerations as for R-fMRI (R-fMRI acquisition strategies section). We will capitalize on the improvements that are identified for R-fMRI early in Phase I by using the same acquisitions for T-fMRI (after confirming with a subset of T-fMRI tasks that the final acquisition protocol works well for task and not just rest). Measures for evaluating acquisition parameters will include assessments of the robustness, spatial extent, and reproducibility of significant task activations and deactivations.
Anatomical MRI acquisition strategies
Conventional structural MRI using T1w scans provide an essential anatomical substrate for visualizing brain structures, generating subcortical segmentations, and reconstructing cortical surfaces. We will also combine anatomical T1w and T2w scans, using the T1w/T2w ratio to map myelin content across the cortical surface and thereby distinguish many architectonic areas non-invasively (Glasser and Van Essen, 2011). This method works with standard 3T 1 mm isotropic T1w and T2w images, but we will explore whether higher resolution images improve architectonic delineations. Additionally at 7T, we will aim to use a similar strategy to map cortical myelin content at 0.6 mm isotropic resolution or higher. Myelin maps will complement other MR modalities in localizing cortical areas in individual subjects and in providing a substrate for improved intersubject registration.
MR scan duration
To obtain the highest quality imaging data feasible for each MR modality, multiple scan sessions are planned for each subject during the 2-day visit. The session structure currently being piloted includes a set of structural scans (20 min total), one diffusion imaging session (1 h), and two 1 h fMRI sessions (each 30 min resting-state followed by 30 min task-fMRI). Participants will be asked if they are willing to undergo an additional voluntary scan session of up to 1 h; this will be used to re-acquire data on any scans that failed to pass initial QC and/or to carry out additional scans using advanced acquisition protocols that might be very informative even if carried out on a modest number of individuals.
MEG/EEG
Non-invasive electrophysiological recording will be carried out in addition to MR scanning and behavioral and genetic testing on 100 subjects (some of whom may also have MR scans at 7T as well as 3T). MEG/EEG is complementary to fMRI in that it provides a window onto the neurophysiological processes underling sensory, motor, and cognitive functions at a temporal scale inaccessible to fMRI. The Blood Oxygen Level Dependent (BOLD) signal detected in fMRI reflects neuronal activity only indirectly; owing to the temporal dynamics of neurovascular coupling (the hemodynamic response function), peak sensitivity to neural activity modulations is on a time scale of seconds (Hathout et al., 1999). In contrast, MEG and EEG respectively detect external magnetic fields and scalp potentials arising from neuronal activity within the brain with millisecond-level temporal resolution. However, the spatial specificity of non-invasive electrophysiology is worse than that of fMRI. Neural sources at the brain surface may be localized with a precision on the order of a few mm, but securely assigning responses to one of multiple simultaneously active generators requires that they be separated by several cm (Mosher et al., 1993). Moreover, MEG sensitivity is largest for parts of the brain within several cm of the sensors; the mesial and inferior cortical surfaces as well as subcortical structures including thalamus and striatum are largely inaccessible. Despite the limited spatial resolution, the richness of temporal information obtained by MEG/EEG enables assessment of how brain rhythmical activity relates to resting and task-evoked connectivity. All these characteristics influence how MEG and EEG data will be integrated with T-fMRI and R-fMRI data, as well as the methods by which cortical parcellation can be applied to these temporally dense signals.
Both R-MEG and T-MEG electrophysiology data will be acquired at SLU using the Magnes 3600 (4D Neuroimaging, San Diego, CA) equipped with 248 magnetometers, 23 MEG reference channels (5 gradiometer, and 18 magnetometer) and 64 EEG Voltage Channels (4 bipolar, 60 monopolar). The system is installed inside a magnetically shielded room that includes one layer of aluminum and two layers of high magnetic permeability material. The RMS noise of the magnetometers is ~5 fT/sqrt (Hz) on average in the white noise range (above 2 Hz). Experience gained during HCP Phase I will determine whether it will be practical to routinely record EEG during Phase II. Prior to MEG/EEG data acquisition, the positions of the EEG electrodes and shape of the subject's head will be mapped by marking fiducials on the subject's skin and using a Polhemus localization system. This will enable co-registration with anatomic MR scans performed subsequently at WashU. The MR data will be used to create anatomic models to support MEG/EEG source reconstruction and will be collected after the MEG/EEG recording session to avoid errors due to subject magnetization. Subjects will complete three resting state scans followed by a set of task runs, with all data collected in a single 2-hour session. MEG/EEG data analyses will be based on the FieldTrip platform (Oostenveld et al., 2011).
The MEG/EEG task paradigms will involve tasks that activate the lateral and dorsal surface of the brain, which are more sensitively sampled by MEG/EEG. In phase I, pilot data will be acquired for motor processes (motor strip mapping), memory (working memory, episodic memory), language, and attention tasks. To facilitate comparisons between T-MEG/EEG and T-fMRI scans, the MEG/EEG task paradigms will be identical in temporal sequence to those used for T-fMRI. Each task under consideration includes sufficient stimuli to allow presentation of different stimuli in each run, thereby avoiding priming effects that might otherwise interfere with subsequent T-fMRI protocols. While the temporal sequence of task protocols will be maintained, T-MEG/EEG protocols may be extended in duration to allow collection of enough trials to ensure adequate sensitivity. Based on the results of these pilot studies, a subset of tasks will be chosen for inclusion in phase II.
A major emphasis of the MEG/EEG component of the HCP will be on developing novel analysis strategies. Non-invasive electrophysiology historically has focused on averaging responses in phase with behaviorally salient events (Dale et al., 2000). Our behavioral protocols will support this methodology but the emphasis will be on analyses of induced oscillatory activity, e.g., event-related time-frequency responses (Hoogenboom et al., 2006) and event-related changes in synchrony within and across brain regions (Siegel et al., 2008). Particular emphasis will be given to novel approaches for analyzing resting state MEG data that require analysis pipelines (Mantini et al., 2011) different from those used for T-MEG paradigms. Patterns of MEG resting connectivity can be studied through e.g., correlation of band-limited power time series (de Pasquale et al., 2010) and characterizing node-pair interactions using complex coherency (Marzetti et al., 2008). Delineation of MEG resting state networks based on beam-former techniques (Brookes et al., 2011) will also be investigated.
Quality assurance
Given the richness and complexity of the datasets to be generated in Phase II of the HCP, it is important to establish and maintain rigorous quality assurance (QA) plans and quality control (QC) processes. Although HCP is a cross-sectional study, the three-year Phase II data collection period and the importance of avoiding drift in ‘healthy normal’ data over time means that many QA and QC challenges faced by longitudinal studies are relevant to HCP. These issues include potential protocol changes, scanner equipment wear, and differences in behavioral interviewing techniques across research staff (Whitney et al., 1998). The HCP Phase II protocols will be fully piloted in late Phase I using adult twins/sibships who do not meet family size criteria for participating in Phase II. We intend that the core HCP protocol, once established, will be invariant throughout Phase II. This protocol will be documented in Standard Operating Procedures made publicly available. Key advances that occur over the course of the study, e.g. in pulse sequences, may be evaluated in additional sessions while the subjects are on-site. To avoid data drift related to equipment performance, scanner QC will be performed daily, and the stability of primary measures associated each data type will be tracked. Many technical aspects of the quality assurance effort are described in Marcus et al. (2011). Efforts to standardize interviewing techniques will include selecting staff to minimize turnover; computerizing the majority of behavioral tests to ensure standard presentation and analysis; and careful training and occasional observation of interviews via audiotapes and two-way mirrors in our testing suite. We will establish an atmosphere in which staff and investigators understand the importance of standardization and are encouraged to discuss and address any issues that might impact this objective.
Discussion
Three issues touched upon above warrant brief discussion. These include issues of (i) limitations of in vivo imaging; (ii) advantages of twin–sibship families coupled with data sharing limitations; and (iii) the relationship of HCP to other large-scale neuroimaging projects.
Inherent limits of in vivo human imaging
Advances in MR scanner design and simultaneous multiplexed data acquisition described above will allow the HCP to generate an unprecedented amount of high quality data on brain connectivity and associated measures in healthy adults (see also Van Essen and Ugurbil, 2012). However, the ‘macro-connectome’ assessments of human brain connectivity accessible via in vivo imaging are on a very different scale than the ‘micro-connectome’ assessments of brain connectivity at the level of single neurons, axons, dendrites, and synapses (Akil et al., 2011). Macro-connectome approaches aim to estimate long-distance connectivity between gray-matter regions using isotropic voxels that are currently often 2 mm (dMRI) or 3 mm (R-fMRI) for 3T and can be 1–2 mm for 7T. The HCP anticipates reducing voxel size for both modalities and for both 3T and 7T, but the scale will remain vastly greater than that of the constituent neuronal elements: human cerebral cortex on average contains ~40,000 neurons and ~3 × 108 synapses per mm,2 and white matter contains ~300,000 axons per mm2 cross-sectional area.3 Micro-connectome approaches are currently restricted to laboratory animals and aim to reconstruct circuitry at scales yet to reach 1 mm3 of brain tissue (Briggman and Denk, 2006; Smith, 2007; Lichtman et al., 2008). Thus, a vast gulf remains between macro- and micro-connectome scales.
Twin–sibship families and data sharing
Our decision to acquire data from twins and non-twin siblings will enable analyses of the heritability of brain circuits and will greatly increase the power of genetic analyses. However, due to the relatively small size and localized geography of the subject population, HCP faces some extra challenges with respect to subject confidentiality and privacy, especially regarding sensitive data. One likely scenario is that the publicly released HCP dataset will include all neuroimaging data and most behavioral data, along with subject sex and age range (e.g., 5-year grouping). Information about family relationships, ethnic and racial identity, exact age (year), and potentially sensitive behavioral measures would be restricted to qualified investigators who agree to appropriate limits on storage and distribution of sensitive data. The publicly released data could also include a dataset consisting of only one individual per family, thereby allowing analyses not confounded by unspecified family relationships.
Relationship to other large-scale imaging projects
A growing number of projects are carrying out large-scale neuroimaging plus behavioral phenotyping on different populations. A non-exhaustive list includes the Alzheimer's Disease Neuroimaging Initiative (ADNI; http://www.adni-info.org/); the Thousand Functional Connectomes project and International Neuroimaging Data-sharing Initiative (INDI, http://fcon_1000.projects.nitrc.org/; Zuo et al., 2010); the IMAGEN study of teenagers and mental health (http://www.imagen-europe.com); the AGES Reykjavik Study of Healthy Aging (http://www.hjarta.is/english/ages); and the Rotterdam study of aging (http://www.epib.nl/research/ergo.htm). Rather than considering each project and associated database as an isolated silo of data, the neuroscience community should make such efforts synergistic to the degree that practical considerations allow. Among the obvious challenges are differences in imaging protocols and scanner hardware, differences in behavioral measures, and different database and data mining platforms. Sharing of information about plans and protocols while there is still flexibility may help to increase commonality in each of these domains and thereby enhance the ability of the community to gain information and insights from data mining that cuts across projects.
In comparison to these other endeavors, the HCP is by no means the largest in terms of the number of subjects studied or in the aggregate amount of data to be collected. However, it is surely the most complex in terms of the diversity of imaging modalities combined with the richness of the behavioral and genetic information to be collected. It also will have an informatics platform that supports an unprecedented degree of visualization and analysis capabilities customized for data mining across all of these modalities. Finally, the HCP is uniquely positioned to improve a variety of data acquisition methods and protocols for brain connectivity studies. An important part of its mission is to openly share these methods as they move from evaluation to production stages. The HCP maintains an active outreach effort to promote awareness in the neuroscience community of the data acquisition strategies outlined here and the informatics strategies described elsewhere (Marcus et al., 2011) and to facilitate coordination with other large-scale neuroimaging projects.
Supplementary Material
Acknowledgments
We thank the entire WU-Minn HCP consortium team for their excellent efforts that have contributed directly or indirectly to the plans described in this manuscript. Funded in part by the Human Connectome Project (1U54MH091657) from the 16 NIH Institutes and Centers that Support the NIH Blueprint for Neuroscience Research, and by the McDonnell Center for Systems Neuroscience at Washington University. Members of the WU-Minn HCP Consortium are listed at http://www.humanconnectome.org/about/hcp-investigators.html and http://www.humanconnectome.org/about/hcp-colleagues.html.
Abbreviations
- ADNI
Alzheimer's Disease Neuroimaging Initiative
- CMRR
Center for Magnetic Resonance Research at UMinn
- dMRI
diffusion imaging
- DZ
dizygotic
- EPI
echoplanar imaging
- HCP
Human Connectome Project, WU-Minn Consortium
- INDI
International Neuroimaging Data-sharing Initiative
- MEG/EEG
magnetoencephalography and electroencephalography
- MZ
monozygotic
- NIRS
near infrared spectroscopy
- PET
positron emission tomography
- R-fMRI
resting state fMRI
- R-MEG
resting state magnetoencephalography
- RF
radio frequency
- SIR
simultaneous image refocusing
- SLU
St. Louis University in St. Louis, MO
- SNR
signal-to-noise ratio
- T-fMRI
task-evoked fMRI
- T-MEG
task-evoked magnetoencephalography
- T1w
T1-weighted
- T2w
T2-weighted
- TE
echo time
- TR
repetition time
- UMinn
University of Minnesota
- WashU
Washington University in St. Louis, MO
- WU-Minn
Washington University and University of Minnesota
- 3T
3 Tesla
- 7T
7 Tesla
Footnotes
Calculations were performed using 3T T2 for white matter, relative to b = 0 for the minimum achievable TE in a Stejskal and Tanner spin echo sequence with one refocusing pulse. Ramp times were ignored for these calculations. The minimum δ (see diagram) was calculated for a given b, G and d (note: Δ = δ + d) by solving 0 = b− (2π·42.58 × 10−3·G·δ)·10−3·(2δ/3 + d) where b is s/mm2, δ and d in ms and G in mT/m. The minimum TE = 2δ + MinTE, where MinTE = minimum TE achievable with δ = 0, d = 0, which was taken to be 15 ms based on existing sequences with partial Fourier acquisition. SNR is calculated using the biexponential diffusion approximation and SNRμ (0.75e−bDF + 0.25e−bDS) e−(2δ+MinTE)/T2 where DF and DS are fast and slow apparent diffusion constants, respectively, (assumed to be 0.8×10−3 and 1×10−4 mm2/s) with corresponding fractional pool sizes of 0.75 and 0.25 (taken from Ronen et al., 2005), with d = 6 ms. White matter 3T T2 was assumed to be 70 ms (Stanisz et al., 2005).
This is based on estimates of 19 billion cortical neurons (Azevedo et al., 2009), 150 trillion cortical synapses (Pakkenberg et al., 2003), and 472 cm3 (4.7 × 104 mm3) cortical gray matter volume (Van Essen et al., 2011).
The human corpus callosum has 2×108 axons (Aboitiz et al., 1992) and a cross-sectional area of 570 mm2 (Rauch and Jinkins, 1996), yielding ~3.5×105 axons per mm2. Human cerebral white matter has a volume of ~700 cm3 (7×105 mm3) (Azevedo et al., 2009; Pakkenberg et al., 2003), and ~150,000 km of aggregate axonal length (150,000–180,000 (Pakkenberg et al., 2003); 120,000 (Tang and Nyengaard, 1997)), for an average of 2.2×105 mm of axonal length per mm3 of white matter.
Appendix A. Supplementary data
Supplementary data to this article can be found online at doi:10.1016/j.neuroimage.2012.02.018.
References
- Aboitiz F, Scheibel AB, Fisher RS, Zaidel E. Fiber composition of the human corpus callosum. Brain Res. 1992;598:143–153. doi: 10.1016/0006-8993(92)90178-c. [DOI] [PubMed] [Google Scholar]
- Achenbach TM, Krukowski RA, Dumenci L, Ivanova MY. Assessment of adult psychopathology: meta-analyses and implications of cross-informant correlations. Psychol. Bull. 2005;131:361–382. doi: 10.1037/0033-2909.131.3.361. [DOI] [PubMed] [Google Scholar]
- Aganj I, Lenglet C, Sapiro G, Yacoub E, Ugurbil K, Harel N. Reconstruction of the orientation distribution function in single- and multiple-shell q-ball imaging within constant solid angle. Magn. Reson. Med. 2010;64:554–566. doi: 10.1002/mrm.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akil H, Martone ME, Van Essen DC. Challenges and opportunities in mining neuroscience data. Science. 2011;331:708–712. doi: 10.1126/science.1199305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S, Skare, Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. NeuroImage. 2003;20:870–888. doi: 10.1016/S1053-8119(03)00336-7. [DOI] [PubMed] [Google Scholar]
- Azevedo FA, Carvalho LR, Grinberg LT, Farfel JM, Ferretti RE, Leite RE, Jacob Filho W, Lent R, Herculano-Houzel S. Equal numbers of neuronal and non-neuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 2009;513:532–541. doi: 10.1002/cne.21974. [DOI] [PubMed] [Google Scholar]
- Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? NeuroImage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggman KL, Denk W. Towards neural circuit reconstruction with volume electron microscopy techniques. Curr. Opin. Neurobiol. 2006;16:562–570. doi: 10.1016/j.conb.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Brookes MJ, Hale JR, Zumer JM, Stevenson CM, Francis ST, Barnes GR, Owen JP, Morris PG, Nagarajan SS. Measuring functional connectivity using MEG: methodology and comparison with fcMRI. NeuroImage. 2011;56:1082–1104. doi: 10.1016/j.neuroimage.2011.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J. Stud. Alcohol. 1994;55:149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Carter AR, Astafiev SV, Lang CE, Connor LT, Rengachary J, Strube MJ, Pope DL, Shulman GL, Corbetta M. Resting interhemispheric functional magnetic resonance imaging connectivity predicts performance after stroke. Ann. Neurol. 2010;67:365–375. doi: 10.1002/ana.21905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. NeuroImage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, Barysheva M, Shattuck DW, Lee AD, Madsen SK, Avedissian C, Klunder AD, Toga AW, McMahon KL, de Zubicaray GI, Wright MJ, Srivastava A, Balov N, Thompson PM. Genetics of brain fiber architecture and intellectual performance. J. Neurosci. 2009;29:2212–2224. doi: 10.1523/JNEUROSCI.4184-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Liu AK, Fischl BR, Buckner RL, Belliveau JW, Lewine JD, Halgren1 E. Dynamic statistical parametric mapping: combining fMRI and MEG for high-resolution imaging of cortical activity. Neuron. 2000;26:55–67. doi: 10.1016/s0896-6273(00)81138-1. [DOI] [PubMed] [Google Scholar]
- de Pasquale F, Della Penna S, Snyder AZ, Lewis C, Mantini D, Marzetti L, Belardinelli P, Ciancetta L, Pizzella V, Romani GL, Corbetta M. Temporal dynamics of spontaneous MEG activity in brain networks. Proc. Natl. Acad. Sci. U. S. A. 2010;107:6040–6045. doi: 10.1073/pnas.0913863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves LJ. The utility of twins. In: Anderon V, et al., editors. Genetic Bases of the Epilepsies. New York: Raven Press; 1982. [Google Scholar]
- Edens EL, Glowinski AL, Pergadia ML, Lessov-Schlaggar CN, Bucholz KK. Nicotine addiction in light smoking African American mothers. J. Addict. Med. 2010;4:55–60. doi: 10.1097/ADM.0b013e3181af111e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estle SJ, Green L, Myerson J, Holt DD. Differential effects of amount on temporal and probability discounting of gains and losses. Mem. Cognit. 2006;34:914–928. doi: 10.3758/bf03193437. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Reese TG, Wedeen VJ. Simultaneous echo refocusing in EPI. Magn. Reson. Med. 2002;48:1–5. doi: 10.1002/mrm.10227. [DOI] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One. 2010;5:e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. Dynamic causal modelling. NeuroImage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC. Myelin content as revealed by T1- and T2-weighted MRI. J. Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn. Reson. Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn. Reson. Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- Gur RC, Richard J, Hughett P, Calkins ME, Macy L, Bilker WB, Brensinger C, Gur RE. A cognitive neuroscience-based computerized battery for efficient measurement of individual differences: standardization and initial construct validation. J. Neurosci. Methods. 2010;187:254–262. doi: 10.1016/j.jneumeth.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Driesen NR, Skudlarski P, Gore JC, Constable RT. Brain connectivity related to working memory performance. J. Neurosci. 2006;26:13338–13343. doi: 10.1523/JNEUROSCI.3408-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathout GM, Gopi RK, Bandettini P, Gambhir SS. The lag of cerebral hemodynamics with rapidly alternating periodic stimulation: modeling for functional MRI. Magn. Reson. Imaging. 1999;17:9–20. doi: 10.1016/s0730-725x(98)00150-7. [DOI] [PubMed] [Google Scholar]
- Hoogenboom N, Schoffelen JM, Oostenveld R, Parkes LM, Fries P. Localizing human visual gamma-band activity in frequency, time and space. NeuroImage. 2006;29:764–773. doi: 10.1016/j.neuroimage.2005.08.043. [DOI] [PubMed] [Google Scholar]
- Larkman DJ, Hajnal JV, Herlihy AH, Coutts GA, Young IR, Ehnholm G. Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. J. Magn. Reson. Imaging. 2001;13:313–317. doi: 10.1002/1522-2586(200102)13:2<313::aid-jmri1045>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Liang ZP, Madore B, Glover GH, Pelc NJ. Fast algorithms for GS-model-based image reconstruction in data-sharing Fourier imaging. IEEE Trans. Med. Imaging. 2003;22:1026–1030. doi: 10.1109/TMI.2003.815896. [DOI] [PubMed] [Google Scholar]
- Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat. Rev. Neurosci. 2008;9:417–422. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D, Della Penna S, Marzetti L, de Pasquale F, Pizzella V, Corbetta M, Romani GL. A signal-processing pipeline for magneto encephalography resting-state networks. Brain Connect. 2011;1:49–59. doi: 10.1089/brain.2011.0001. [DOI] [PubMed] [Google Scholar]
- Marcus D, Harwell J, Olsen T, Van Essen D. The Human Connectome Project informatics platform. Frontiers in Neuroscience. 2011;5:4. doi: 10.3389/fninf.2011.00004. (Electronic publication ahead of print 2011 Jun 27). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Boomsma D, Machin G. A twin-pronged attack on complex traits. Nat. Genet. 1997;17:387–392. doi: 10.1038/ng1297-387. [DOI] [PubMed] [Google Scholar]
- Marzetti L, Del Gratta C, Nolte G. Understanding brain connectivity from EEG data by identifying systems composed of interacting sources. NeuroImage. 2008;42:87–98. doi: 10.1016/j.neuroimage.2008.04.250. [DOI] [PubMed] [Google Scholar]
- McCrae RR, Costa PT., Jr A contemplated revision of the NEO Five Factor Inventory. Personal. Individ. Differ. 2004;36:587–596. [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 Tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson. Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher JC, Spencer ME, Leahy RM, Lewis PS. Error bounds for EEG and MEG dipole source localization. Electroencephalogr. Clin. Neurophysiol. 1993;86:303–321. doi: 10.1016/0013-4694(93)90043-u. [DOI] [PubMed] [Google Scholar]
- Neale MC, Cardon LRE. Methodology for Genetic Studies of Twins and Families. Dordrecht: Kluwer Academic Publishers; 1992. [Google Scholar]
- Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, Velanova K, Donaldson DI, Phillips JS, Schlaggar BL, Petersen SE. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010;67:156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oostenveld R, Fries P, Maris E, Schoffelen JM. FieldTrip: open source software for advanced analysis of MEG, EEG, and invasive electrophysiological data. Comput. Intell. Neurosci. 2011 doi: 10.1155/2011/156869. 156869 EPub 152010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakkenberg B, Pelvig D, Marner L, Bundgaard MJ, Gundersen HJ, Nyengaard JR, Regeur L. Aging and the human neocortex. Exp. Gerontol. 2003;38:95–99. doi: 10.1016/s0531-5565(02)00151-1. [DOI] [PubMed] [Google Scholar]
- Posthuma D, Boomsma DI. A note on the statistical power in extended twin designs. Behav. Genet. 2000;30:147–158. doi: 10.1023/a:1001959306025. [DOI] [PubMed] [Google Scholar]
- Power J, Barnes K, Snyder A, Schlaggar B, Petersen S. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn. Reson. Med. 1999;42:952–962. [PubMed] [Google Scholar]
- Rauch RA, Jinkins JR. Variability of corpus callosal area measurements from midsagittal MR images: effect of subject placement within the scanner. AJNR Am. J. Neuroradiol. 1996;17:27–28. [PMC free article] [PubMed] [Google Scholar]
- Ronen I, Ugurbil K, Kim DS. How does DWI correlate with white matter structures? Magn. Reson. Med. 2005;54:317–323. doi: 10.1002/mrm.20542. [DOI] [PubMed] [Google Scholar]
- Sartor CE, Bucholz KK, Nelson EC, Madden PA, Lynskey MT, Heath AC. Reporting bias in the association between age at first alcohol use and heavy episodic drinking. Alcohol. Clin. Exp. Res. 2011;35:1418–1425. doi: 10.1111/j.1530-0277.2011.01477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Chang S, Hoyer P, Hyvarinen A, Kerminen A. A linear non-Gaussian acyclic model for causal discovery. J. Mach. Learn. Res. 2006;7:2003–2030. [Google Scholar]
- Siegel M, Donner TH, Oostenveld R, Fries P, Engel AK. Neuronal synchronization along the dorsal visual pathway reflects the focus of spatial attention. Neuron. 2008;60:709–719. doi: 10.1016/j.neuron.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Smith SJ. Circuit reconstruction tools today. Curr. Opin. Neurobiol. 2007;17:601–608. doi: 10.1016/j.conb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain's functional architecture during activation and rest. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for FMRI. Neuro-Image. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Smith S, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Van Essen D, Feinberg D, Yacoub E, Ugurbil K. Temporally-independent functional modes of spontaneous brain activity. Proc. Natl. Acad. Sci. 2012;109:3131–3136. doi: 10.1073/pnas.1121329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodickson DK, Griswold MA, Jakob PM. SMASH imaging. Magn. Reson. Imaging Clin. N. Am. 1999;7:237–254. vii–viii. [PubMed] [Google Scholar]
- Stanisz GJ, Odrobina EE, Pun J, Escaravage M, Graham SJ, Bronskill MJ, Henkelman RM. T1, T2 relaxation and magnetization transfer in tissue at 3T. Magn. Reson. Med. 2005;54:507–512. doi: 10.1002/mrm.20605. [DOI] [PubMed] [Google Scholar]
- Tang Y, Nyengaard JR. A stereological method for estimating the total length and size of myelin fibers in human brain white matter. J. Neurosci. Methods. 1997;73:193–200. doi: 10.1016/s0165-0270(97)02228-0. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Gadian DG, Connelly A. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage. 2004;23:1176–1185. doi: 10.1016/j.neuroimage.2004.07.037. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Kahn RS, Hulshoff Pol HE. Efficiency of functional brain networks and intellectual performance. J. Neurosci. 2009;29:7619–7624. doi: 10.1523/JNEUROSCI.1443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K. The future of the human connectome. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2012.01.032. http://dx.doi.org/10.1016/j.neuroimage.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Glasser MF, Dierker D, Harwell J, Coalson T. Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb. Cortex. 2011 doi: 10.1093/cercor/bhr291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, Pandya DN, Hagmann P, D'Arceuil H, de Crespigny AJ. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. NeuroImage. 2008;41:1267–1277. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Whitney CW, Lind BK, Wahl PW. Quality assurance and quality control in longitudinal studies. Epidemiol. Rev. 1998;20:71–80. doi: 10.1093/oxfordjournals.epirev.a017973. [DOI] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Di Martino A, Mennes M, Margulies DS, Bangaru S, Grzadzinski R, Evans AC, Zang YF, Castellanos FX, Milham MP. Growing together and growing apart: regional and sex differences in the lifespan developmental trajectories of functional homotopy. J. Neurosci. 2010;30:15034–15043. doi: 10.1523/JNEUROSCI.2612-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.