examples
brat can be applied in many ways and is used in several completed and ongoing annotation efforts and other tasks:
annotation examples in brat
A variety of annotation tasks that can be performed in brat are introduced below using examples from available corpora. The examples discussed in this section have been originally created in various tools other than brat and converted into brat format. Converters for many of the original formats are distributed with brat. In the selection of examples included here, priority has been given to tasks with freely available data.
Entity mention detection

Example: CoNLL 2002 Shared Task: Language-Independent Named Entity Recognition
The Conference on Computational Natural Language Learning (CoNLL) 2002 shared task on Language-Independent Named Entity Recognition provided two annotated corpora (Spanish and Dutch) annotated with entities of four types (person, organization, location and miscellaneous).
A conversion script from the CoNLL 2002 shared task format into the brat standoff format and a sample of the corpus annotations are distributed with brat.
The full shared task data are freely available from the shared task website.
Event extraction

Example: BioNLP shared task: biomedical event extraction
BioNLP shared task events held in 2009 and 2011 and the ongoing BioNLP Shared Task 2013 a have included several event extraction tasks.
The brat standoff format is compatible with the data distribution format of the BioNLP shared task, and samples of the annotations of the various corpora provided for the task are distributed with brat.
The full shared task data are freely available from the shared task website.
Coreference resolution

Example: CO supporting task: coreference in scientific publications
The BioNLP shared task 2011 included a supporting task on coreference in scientific publications.
The brat standoff format is compatible with the representation used in the coreference task, and samples of the annotations of the corpus provided for the task are distributed with brat.
The full shared task data are freely available from the shared task website.
Normalization

Example: the Colorado Richly Annotated Full Text Corpus (CRAFT)
The Colorado Richly Annotated Full Text Corpus (CRAFT) is an annotated corpus of full text biomedical journal publications. Mentions of concepts from seven biomedical ontologies/ terminologies, including ChEBI, GO, and the NCBI Taxonomy are annotated and normalized to the appropriate ontology identifier.
The corpus creators have converted the annotation into the brat format and provide an online service for browsing the CRAFT concept annotations in brat.
The full corpus is freely available from the project homepage.
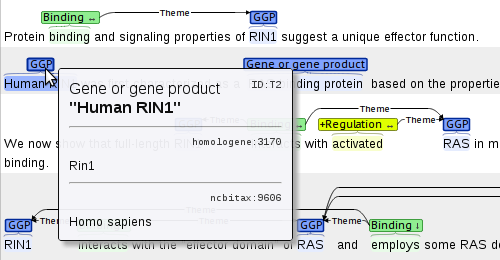
Example: EVEX: large-scale event extraction resource with multi-level gene normalization
The EVEX database builds on an automatically annotated corpus covering 22 million PubMed abstracts and 460 thousand open access full-text articles from PubMed Central, created using tools such as the Turku Event Extraction System and the gene normalization system GenNorm. Each gene and protein mention in the data is normalized to several databases at varying levels of granularity, ranging from canonical symbols to broad gene families and unique Entrez Gene IDs.
The corpus creators have converted the annotation into the brat format. A (very) small sample is linked below.
The full corpus is freely available from bionlp.utu.fi/pubmedevents.html.
Chunking
Chunking is the task of dividing text into non-overlapping segments that are typically further assigned labels such as NP (Noun Phrase).

Example: CoNLL 2000 Shared Task: Chunking
The Conference on Computational Natural Language Learning 2000 (CoNLL 2000) shared task on chunking provides freely available training and test data.
A conversion script from the CoNLL 2000 shared task format into the brat standoff format and a sample of the corpus annotations are distributed with brat.
The full shared task data are freely available from the shared task website.
Dependency syntax
Dependency parsing (syntactic analysis) is the task of assigning binary relations between words to mark their head-dependent relations.

Example: CoNLL-X Shared Task: Multi-lingual Dependency Parsing
The Tenth Conference on Computational Natural Language Learning (CoNLL-X) shared task on Multi-lingual Dependency Parsing provided annotated corpora for 13 languages, four of which are freely availabe (for Danish, Dutch, Portuguese and Swedish).
A conversion script from the CoNLL-X shared task format into the brat standoff format and a sample of the corpus annotations for these four languages are distributed with brat.
The full shared task data are freely available from the shared task website.

Example: Turku Dependency Treebank (TDT)
TDT is a Finnish treebank manually annotated using the Stanford Dependencies (SD) scheme. In addition to the base syntactic trees, TDT also includes additional relations, such as coordination expansion. The underlying corpus comprises a number of different genres, including among others legalese, encyclopedic text, fiction, and news. A conversion script from TDT's native XML format into the brat standoff format is available upon request and a sample of the treebank is distributed with brat.
Meta-knowledge
Meta-knowledge annotation is the task of identifying how factual statements should be interpreted, according to their textual context, Examples include whether a statement describing a fact, a hypothesis, an experimental result or an analysis of results, how confident the author is regarding the validity of her analyses, etc.

Example: Meta-knowledge enrichment of the GENIA Event Corpus
The complete GENIA event copus, consisting of 1,000 abstracts annotated with 36,858 events, has been manually enriched with meta-knowledge annotation. This will faciltate the training of more sophisticated IE systems, which allow interpretative information to be specified as part of the search criteria. Such systems can assist in a number of important tasks, e.g., finding new experimental knowledge to facilitate database curation, enabling textual inference to detect entailments and contradictions, etc.
Each event has been annotated with 5 different dimensions of meta-knowledge, i.e. knowledge type (KT), certainty level (CL), manner, polarity and source. The annotation consists of assigning an appopriate value for each dimension and identifying textual clues for each dimension, when they are present.
The meta-knowledge enririched GENIA corpus is freely available from the project website.
corpora annotated using brat
This section presents a selection of corpora annotated using brat.
Information Extraction
Information Extraction is the task of capturing essential information contained in text and convert it into a formal representation suitable for automatic processing.

Anatomical Entity Mention (AnEM) corpus
brat was used to create AnEM, a corpus of 500 documents drawn from PubMed abstracts and PMC full text extracts, annotated for 11 classes of anatomical entities. The corpus is intended to serve as a reference for training and evaluating methods for anatomical entity mention detection in life science publications.

CellFinder corpus
brat was used to annotate the CellFinder corpus of 10 full papers annotated with six biomedical entities: genes/proteins, cell components, cell types, cell lines, anatomical parts (tissues/organs) and organisms. The corpus was developed in the scope of the CellFinder project which aims to establish a central stem cell data repository.
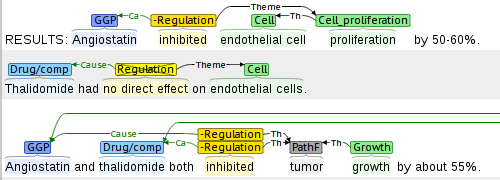
Multi-Level Event Extraction (MLEE) corpus
brat was used to create the Multi-Level Event Extraction (MLEE) corpus of 260 PubMed abstracts annotated with 16 physical biomedical domain entity types ranging from molecular entities to cells, tissues, and organs and 29 event types involving these entities, defined primarily with reference to the Gene Ontology.
The corpus is currently being applied and extended in the BioNLP Shared Task 2013 Cancer Genetics task.
Metaphor annotation through bottom-up identification
Bottom-up metaphor annotation is the task of identifying linguistic metaphors in the text and inferring the conceptual metaphors that underlie them.
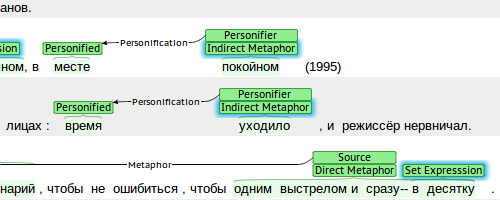
Russian-language corpus of conceptual metaphor
The manually annotated Russian-language corpus of conceptual metaphor is an ongoing project in the initial stage. The annotation is performed at the two levels. At the shallow level, metaphor-related words are annotated according to slightly modified rules of MIPVU, the linguistic procedure of metaphor identification. Each metaphor-related word is assigned to one of the 10 classes. The word meanings that form the basis of metaphoricity are described for each metaphor-related word.
The deep-level annotation consists in annotating metaphorical mappings between the conceptual domains of the Source and the Target, and inferring conceptual metaphors from them.
This scheme is suitable for identifying and annotating conceptual metaphors that are partly manifested in the text, with only the Source being present explicitly, while the Target and the mapping are implicit.
other uses of brat
brat can be used also for tasks other than manual annotation.
Visualisation
brat has not just been used to create annotations but also to visualise annotation output from other tools for manual analysis and evaluation purposes. Instructions on how to embed brat visualisations in web pages are available here.

Natural language analysis tool visualization
The Standford NLP group is using brat to provide visualizations of the annotations created by the live online demo of the Stanford CoreNLP natural language analysis tools.
The CoreNLP tools perform part-of-speech tagging, named entity recognition, coreference resolution, and syntactic analysis. The demonstration provides separate visualizations of all of these annotation layers using embedded brat.
Information Extraction system evaluation

Example: BioNLP shared task: biomedical event extraction
BioNLP Shared Task 2011 and the ongoing BioNLP Shared Task 2013 are using brat to provide visualizations of the results of information extraction system predictions compared to gold standard annotations.
The evaluation system evaluates differences between two sets of annotations and generates a visual comparison in brat format, using brat features to make differences visually obvious.
