Abstract
Moso bamboo (Phyllostachys edulis), an ecologically and economically important forest species in East Asia, plays vital roles in carbon sequestration and climate change mitigation. However, intensifying climate change threatens moso bamboo survival. Here we generate high-quality haplotype-based pangenome assemblies for 16 representative moso bamboo accessions and integrated these assemblies with 427 previously resequenced accessions. Characterization of the haplotype-based pangenome reveals extensive genetic variation, predominantly between haplotypes rather than within accessions. Many genes with allele-specific expression patterns are implicated in climate responses. Integrating spatiotemporal climate data reveals more than 1050 variations associated with pivotal climate factors, including temperature and precipitation. Climate-associated variations enable the prediction of increased genetic risk across the northern and western regions of China under future emissions scenarios, underscoring the threats posed by rising temperatures. Our integrated haplotype-based pangenome elucidates moso bambooâs local climate adaptation mechanisms and provides critical genomic resources for addressing intensifying climate pressures on this essential bamboo. More broadly, this study demonstrates the power of long-read sequencing in dissecting adaptive traits in climate-sensitive species, advancing evolutionary knowledge to support conservation.
Similar content being viewed by others
Introduction
Moso bamboo (Phyllostachys edulis) is an ecologically and economically vital forest species that is distributed extensively across East Asia and plays crucial roles in carbon sequestration and climate change mitigation1. The Chinese bamboo market, valued at $41.58 billion in 2020, is projected to surpass $138.63 billion by 20352,3, underscoring the increasing economic significance of the bamboo industry worldwide and its potential impact on the global market. With its rapid growth and high carbon storage capacity, one hectare of moso bamboo forest can store twice as much carbon as Chinese fir forests over a 60-year period4. However, intensified climate change seriously threatens plant biodiversity and survival, which will hinder the reduction of greenhouse gases via carbon sinks and affect the Paris Agreement5,6,7. For habitat-bound species such as moso bamboo, the continuous accumulation of genetic variation is crucial for the generation of adaptable individuals who can respond to climate change8,9. A previous study indicated that moso bamboo populations have undergone two bottleneck events, resulting in a low genetic diversity of 0.02710. This lack of diversity, coupled with the inherent climate sensitivity of moso bamboo11,12, highlights the risk to its survival in the face of intensified climate change. Therefore, a quantitative assessment of the climate risks and adaptation capacity of moso bamboo would benefit protection policies and breeding efforts; however, such studies remain scarce.
In recent years, substantial progress has been made in evaluating plant population vulnerability to climate change through genomic approaches13. Several studies on forest tree species have demonstrated the efficacious identification of climate-adaptive variations and the ability to predict climate responses by integrating single-nucleotide polymorphism (SNP) and environmental data to detect their inherent associations14,15. Genomic offset represents the disruption of current genotypeâclimate relationships due to rapid shifts in climate16,17. The advantage of genomic offset approaches lies in their ability to predict population responses and vulnerability to climate change from genomic data, serving as an alternative to common garden experiments18,19. Forward and reverse genomic offsets can also predict population maladaptation under migration scenarios13. However, SNPs may not fully capture climate-associated genomic information since other types of genetic variations, such as insertions and deletions (InDels) and structural variations (SVs) are usually ignored14. Furthermore, systematic and accurate detection of SVs and allele-specific expression (ASE) is challenging due to limitations in sequence resolution, although they play important roles in speciation and adaptive evolution in plants20. These limitations in detecting genetic variations hinder the identification of loci associated with climate adaptation. Therefore, haplotype genome sequencing enables higher-resolution localization of genetic variations, expediting the dissection of these complex genomic traits and improving the understanding of genetic diversity, genotypeâphenotype associations, and environmental adaptation in tree populations21.
Recent studies have also shown the power of pangenome analysis for comprehensively elucidating genetic diversity by integrating de novo genome assemblies of multiple accessions22. However, pangenome analysis still relies on short-read sequencing, which has limited power in systematically capturing SVs compared to long-read sequencing technologies. Although pangenome analysis is becoming increasingly accessible, investigations into long-read haplotypes and SVs remain scarce for nonmodel species23, such as moso bamboo. Therefore, constructing high-quality haplotype-based pangenomes using long-read sequencing will enable more comprehensive identification of genetic variations associated with adaptation in moso bamboo.
In this study, we construct a haplotype-based pangenome for moso bamboo using PacBio HiFi and Hi-C sequencing strategies to elucidate the genetic basis underlying the wide distribution and climate adaptation of this species. By integrating comprehensive genomic datasets from 16 representative moso bamboo accessions (RMAs), we characterize genome-wide genetic variations and ASE at high resolution. Furthermore, by leveraging a graph-based pangenome and high-resolution spatiotemporal climate data, we identify genetic loci associated with local climate adaptation and quantify climate maladaptation risk across moso bamboo populations in China. Our research addresses the following key questions: 1) What is the extent and pattern of the haplotype-level genomic diversity of moso bamboo? 2) How does ASE contribute to the adaptive resilience of moso bamboo? 3) Which genomic variations underlie local climate adaptation in moso bamboo populations? 4) How will projected climate change scenarios impact the climate maladaptation risks of moso bamboo populations? Addressing these questions provides critical insights to inform evidence-based conservation and breeding strategies for safeguarding this ecologically and economically vital species in the face of rapid global climate change.
Results
Genome sequencing and variation identification of 16 representative moso bamboo accessions
We selected 16 representative moso bamboo accessions (RMAs) according to their geographical distribution across China to capture extensive genetic diversity (Supplementary Fig. 1). The genomes of all the RMAs were de novo sequenced, generating 1.03 Tb of PacBio HiFi reads, resulting in an average sequencing depth of 33.74à per RMA (Supplementary Table 1). These data allowed the construction of 16 high-quality assemblies (32 haplotype assemblies) with an average contig N50 length of 57.0âMb (Table 1 and Supplementary Fig. 2). The average quality value (QV) of the final assembly was 64.26, with a k-mer completeness of 98.20% (Supplementary Fig. 3). We observed an average switch error of 5.44% for all assemblies (Table 1 and Supplementary Table 2). Additionally, we employed Hi-C sequencing to facilitate chromosome-level genome assembly for three RMAs (CY, HB, and HZP), yielding an average sequencing depth of 139.24à (Supplementary Table 3). Hi-C interactions were utilized to anchor, order, orient, and correct contig misjoins, enabling 99.07% of the sequences to be anchored to pseudochromosomes across haplotype assemblies (Table 1).
To identify high-quality protein-coding gene models, we sequenced the transcriptomes of 3 or 4 tissues from each RMA, generating 1.34âTb of RNA-seq data from 186 samples (Supplementary Data 1). Using a combinatorial approach (see Methods), we predicted an average of 54,343 protein-coding gene models in all 32 haplotype assemblies (Table 1), with an average of 97.9% of them assigned putative functions (Supplementary Data 2). Among these gene models, an average of 92,506 genes were present as biallelic genes (corresponding to 46,253 allele pairs in each RMA), while 8090 genes, on average, were present as single alleles in the haplotype assemblies (Supplementary Table 4). The genome assemblies and annotations were subjected to extensive quality evaluations, including assessments of continuity, base accuracy, structural accuracy, and correlation analyses, which revealed high-quality haplotype assemblies and gene models suitable for downstream analyses (Supplementary Data 3 and Supplementary Fig. 4). We also identified intact long terminal repeats (LTRs) across the 16 assemblies and clustered the LTR families (Supplementary Figs. 5â6).
For the comprehensive characterization of the genetic variations across the 16 RMAs, we selected CYhap1 as the reference genome due to its superior quality compared to the other accessions, and successfully resolved its haplotypes. Thus, we identified 2123,198 SNPs and 294,108 InDels (Fig. 1a), collectively referred to as short variations in this study. We found that over half (65.9%) of the short variations were present in all 16 accessions or only one accession (Supplementary Fig. 7). Additionally, we detected 26,987 SVs, including 12,417 insertions (INSs), 14,494 deletions (DELs), and 76 inversions (INVs), using five SV callers based on two strategies (Fig. 1a, Supplementary Data 4 and Supplementary Fig. 8). Similarly, 63.8% of the SVs were shared by all 16 accessions or only one accession (Supplementary Fig. 7). Subsequently, we utilized these SVs to construct a graph-based pangenome. Furthermore, we identified SVs intersecting with LTRs, and the formation of these SVs might be associated with the presence of LTRs (Supplementary Data 5).
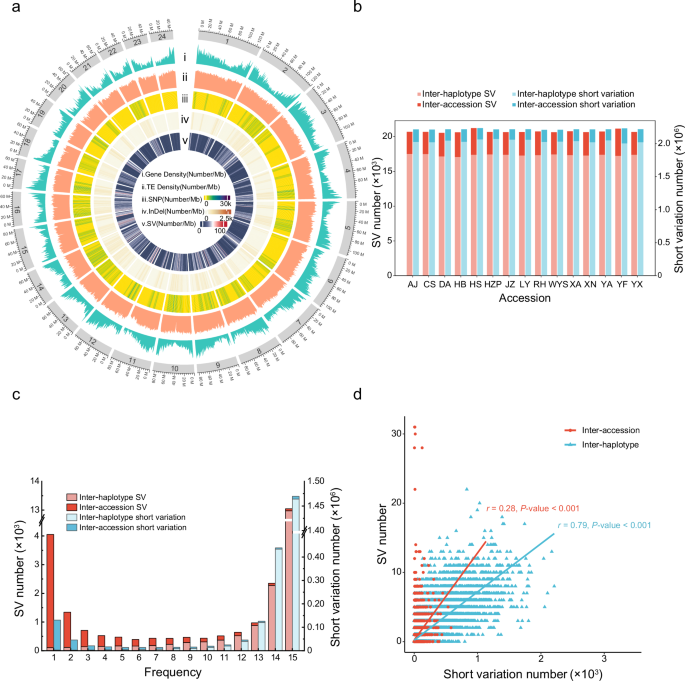
a Circle plot visualizing the moso bamboo pangenome, with concentric tracks showing the densities of genes, transposable elements (TEs), single-nucleotide polymorphisms (SNPs), small insertions and deletions (InDels), and structural variations (SVs) from the outermost to innermost rings (from i to v). b Numbers of SVs (red) and short variations (SNPs and InDels, blue) categorized as either inter-accession (darker colors) or inter-haplotype (lighter colors). Inter-accession variations are absent between the reference haplotypes but are present in other accessions. Inter-haplotype variations are present between the reference haplotypes. The x-axis represents accessions, and the y-axis shows the number of SVs/short variations. c Frequency distributions (x-axis) of inter-haplotype and inter-accession SVs/short variations (y-axis). d Correlation between the genomic distributions of SVs and short variations per 100âkb genomic window. Inter-haplotype variations (red) show stronger correlations (two-sided Pearsonâs correlation test with P-valueâ=â0â<â0.001) in their genome-wide distributions than inter-accession variations (blue, two-sided Pearsonâs correlation test with P-valueâ=â0â<â0.001).
Most of the genetic variation in moso bamboo occurred between haplotypes rather than within accessions
By comparing the genetic differences of each RMA with the variations between the haplotypes of the reference genome, we found that, on average, 97.0% of the heterozygous variations were also present between the haplotypes of the reference genome. Thus, these variations were classified as being present between the haplotypes of each accession (termed âinter-haplotypeâ in this study), rather than between the accessions (referred to as âinter-accessionâ). The numbers of inter-haplotype short variations and SVs were on average 10.4 times and 5.3 times greater, respectively, than those of inter-accession short variations and SVs (Fig. 1b and Supplementary Table 5). The average levels of heterozygosity of total short variations and inter-accession short variations were 98.6% and 71.2%, respectively (Supplementary Data 6), suggesting that traditional methods of variant identification overestimate heterozygosity in moso bamboo. On average, we identified one short variation per 923.9âbp and one inter-accession short variation per 23,105.9âbp (Supplementary Table 6), suggesting that the actual genetic diversity in moso bamboo was lower than that previously reported10.
Among all the RMAs, for short variations and SVs, 68.5% and 68.8% of the inter-haplotype variations were highly frequent (detected in all the accessions), whereas only 0.1% and 0.5%, respectively, were less frequent (detected in only one accession). In contrast, most inter-accession variations (46.3% of short variations and 49.7% of SVs) were present at low frequencies, whereas only 0.2% and 0.9%, respectively, had high frequencies (Fig. 1c). Furthermore, we compared the three well-phased genomic haplotypes and calculated the number of SNPs, revealing that the differences among the same haplotypes from different accessions were relatively small, while the variations between different haplotypes were larger (Supplementary Table 7). Therefore, the results revealed that the accessions harbored largely similar inter-haplotype variations, substantially exceeding the inter-accession variations. The standard deviation revealed that inter-haplotype variations also represented more clustered genomic distributions than did inter-accession variations (Supplementary Table 8 and Supplementary Fig. 9). We also observed a strong positive correlation (Pearsonâs râ=â0.79) between genome-wide distributions of short variations and SVs for inter-haplotype variations compared with inter-accession variations, which showed a weak positive correlation (Pearsonâs râ=â0.28) (Fig. 1d). With respect to the length of the SVs, those lengths less than 100âkb had more inter-haplotype SVs, while 95% of the SVs with lengths greater than 100âkb were inter-accession SVs (Supplementary Fig. 10).
Construction and characterization of a haplotype-based pangenome for moso bamboo via allele comparison
To better characterize haplotypes and comparatively analyze alleles, we aligned and identified alleles based on protein sequence similarity and intergenic distance (see Methods). Thus, a haplotype-based pangenome for moso bamboo was constructed by identifying and comparing alleles across RMAs. In total, 1738,962 genes were grouped into 74,758 gene sets from the 32 haplotype assemblies. Gene sets were categorized by their presence across accessions as core (present in all 16 accessions), softcore (present in 12â15 accessions), dispensable (present in 2â11 accessions), or private (present in only one accession) gene sets. The proportions of these four categories were 53.90%, 16.94%, 28.06%, and 1.10%, respectively (Fig. 2b). Additionally, the gene sets were divided into three groups based on allele composition: double-allele gene sets (the allele pair was detected in all accessions), single-allele gene sets (only one allele of each allele pair was detected in all accessions), and variable-allele gene sets (the allele pair was detected in some accessions). A schematic diagram illustrating the classification is provided in Supplementary Fig. 11. Among the 12 groups resulting from the combination of gene frequency and allele composition categories, core gene sets accounted for the greatest proportion (92.1%) of the double-allele gene sets, while they accounted for the lowest proportion (0.3%) of the single-allele gene sets (Fig. 2c).
a The number of gene sets in the pangenome (blue) and core gene set (red) increased as a function of the number of moso bamboo accessions included in the analysis (x-axis). The error bars represent the mean valuesâ±âSDs, n = simulation times. b Compositions of the pangenome. The bar plots show the number of gene sets (y-axis) in each accession categorized by frequency (x-axis). The pie chart depicts the proportions of gene sets marked by each composition category: core, softcore, dispensable, and private gene sets. The left block shows the number of unique gene sets (bottom) and the sum of unclustered genes in each genome (hatched area). c Distribution of gene sets in different groups based on gene frequency and allele composition. The y-axis represents the four groups divided according to gene frequency across accessions, and the x-axis represents the 3 gene set groups categorized according to allele composition. All the gene sets were divided into 12 groups (core-double (19,270), core-variable (20,858), core-single (169), softcore-double (561), softcore-variable (11,772), softcore-single (330), dispensable-double (267), dispensable-variable (17,010), dispensable-single (3,702), private-double (819), private-variable (5998), and private-single (44,104)). The area of each group is proportional to the number of gene sets. d, e, and f Comparison of gene length (d), expression level (e), and tissue specificity index (Tau) (f) across the 12 gene set groups (x-axis). The y-axes show gene length in base pairs, log2(TPMâ+â1) expression values, and the Tau specificity index. The box plots show the medians (centerlines), interquartile ranges (boxes), and 1.5 times the interquartile ranges (whiskers). n = gene set numbers. The P-values indicating statistical significance are provided in Supplementary Tables 9â11.
Characterization of the 12 gene set groups revealed differences in gene structure, expression pattern, and functional features. The gene length, cDNA length, coding DNA sequence (CDS) number, and CDS size in the core gene sets were greater than those in the private gene sets within the same haplotype category, and those in the double-allele gene sets were greater than those in the single-allele gene sets (Wilcoxon signed-rank test, P-valueâ<â0.001) (Fig. 2d, Supplementary Figs. 12â14 and Supplementary Table 9). The average gene expression level (transcripts per kilobase million, [TPM]) gradually decreased from the core to the private gene sets and decreased from the double-allele to single-allele gene sets, except for the private gene sets (Fig. 2d and Supplementary Table 10). The tissue specificity index (Tau) was significantly lower in the core gene sets than in the private gene sets (Wilcoxon signed-rank test, P-valueâ<â0.001), and the Tau in the single-allele gene sets was greater than that in the double-allele gene sets, except for the private gene sets (Wilcoxon signed-rank test, P-valueâ<â0.05) (Fig. 2e and Supplementary Table 11). These results showed that more genes were expressed and that their expression levels were greater in the core gene sets, whereas those in the private gene sets exhibited greater tissue specificity. A similar pattern was observed in the double-allele and single-allele gene sets. Additionally, we focused on the core-single gene set, which represents genes present in all the accessions but only in one haplotype assembly. Among the 47 core-single gene sets with known functions and a TPM greater than 1 in at least one RNA-seq accession, 27 gene sets whose functions were related to environmental adaptation were identified (Supplementary Data 7). The functions of these gene sets include stress tolerance (e.g., the gene set GS0035370, encoding aldo-keto reductase 1 (AKR1))24, disease resistance (e.g., the gene set GS0058418, encoding disease resistance protein RPM1)25, and DNA damage repair (e.g., the gene set GS0062031, encoding dynamics of the (6-4) photolyase)26. These results suggested that some haplotype-specific genes may be involved in the environmental adaptation of moso bamboo.
Involvement of allele-specific gene expression in the environmental adaptation of moso bamboo
Allele-specific expression (ASE) analysis revealed 16,317 genes exhibiting ASE across the 16 pairs of haplotype assemblies (Supplementary Data 8). These genes were distributed across 8730 gene sets in the pangenome, with the largest proportion (34.1%) belonging to the core-variable gene set and the fewest (0.3%) belonging to the dispensable-double gene set (Fig. 3a). Additionally, the distribution of allele-specific expression gene sets (ASEGs) revealed a high degree of accession specificity, with 81.8% (7139) of ASEGs detected in only 1â2 accessions and only 9 ASEGs shared among all accessions (Fig. 3b). Interestingly, ASEGs from a common gene set also exhibited variable allele-specific expression patterns among tissues. For instance, of the 3149 ASEGs detected in two or more accessions across different haplotype assemblies, 72% exhibited tissue-specific expression patterns that varied between accessions, while the remaining 28% demonstrated consistent tissue expression across accessions (Supplementary Data 9).
a Distribution of allele-specific expression gene sets (ASEGs) across the moso bamboo pangenome categorized by accession (inner circle) and allele (outer circle). b Frequency distribution of ASEGs (y-axis) across all accessions (x-axis). c Tissue-specific distribution of ASEGs detected in all four tissues (leaf, stem, rhizome, and root) in the moso bamboo pangenome. d Gene Ontology (GO) enrichment analysis of ASEGs in all four tissues and exclusively in one tissue using a one-sided hypergeometric distribution test. The circle color represents the statistical significance (P-value), and the size represents the number of ASEGs. The rich factor is the ratio of ASEGs annotated to a given GO term over the total number of genes in that term. e An example of a consistent ASEG (the gene set GS0010347) exhibiting ASE patterns. The ASEG showed high expression (red) in haplotype 1 and low expression (blue) in haplotype 2 in both the stem and rhizome. f Schematic representation of a 6398-bp SV in GS0010347 that may induce ASE by altering protein sequences between haplotypes. The gene regions are green, the CDSs are blue, and the 5â/3â UTRs are red. g An example of an inconsistent ASEG (the gene set GS0027844) exhibiting tissue-specific patterns. For the five accessions (AJ, CY, RH, XA, and XN), the ASEGs showed greater expression in haplotype 1 than in haplotype 2 in the leaf. Conversely, in the rhizome, the same ASEGs exhibited lower expression in haplotype 1 and higher expression in haplotype 2 relative to the leaf pattern. h Schematic representation of a DEL and several base substitutions between the CDSs of the two haplotypes in the gene set GS0027844. These sequence differences (red) changed the protein sequence between the two haplotypes.
ASE tissue-specific analyses revealed that the highest proportion of ASEGs (39.3%) were expressed in all tissues, followed by ASEGs expressed exclusively in leaves (13.4%), roots (6.1%), stems (5.1%), and rhizomes (4.9%) (Fig. 3c). Functional enrichment analysis of these ASEGs revealed associations with various stimuli and defense responses (Fig. 3d). ASEGs expressed in all tissues were primarily enriched for processes related to protein biosynthesis and modification, whereas those with tissue-specific expression were involved in developmental processes, such as wax biosynthetic processes in leaves and cell wall biogenesis in stems (Fig. 3d). These results indicated that moso bamboo ASEGs likely play key roles in environmental adaptation while also contributing to tissue-specific developmental processes.
Subsequent analysis revealed 5156 consistent ASEGs (exhibiting consistent expression patterns toward one allele across tissues) derived from 3183 gene sets (Supplementary Data 10). For example, the gene set GS0010347 showed consistent ASEGs across all the accessions in the stems and rhizomes, representing a ubiquitous ASEG among the haplotype assemblies (Fig. 3e). The CDS of GS0010347 overlapped with a 6398-bp inter-haplotype DEL (Fig. 3f), potentially resulting in ASE. This gene set encodes flavin-containing monooxygenase 1 (FMO1), which has been experimentally shown to participate in establishing systemic acquired resistance27. These results suggest that certain inter-haplotype variations could be associated with consistent ASE events. In addition, we identified 68 inconsistent ASEGs (exhibiting high expression switching between alleles in different tissues) derived from 47 gene sets (Supplementary Data 11). For instance, the gene set GS0027844 exhibited leaf ASE in 5 accessions (AJ, CY, RH, XA, and XN) with high expression in haplotype 1 but showed the opposite pattern in the rhizome, with high expression in haplotype 2 (Fig. 3g). The ASE was potentially induced by a 6-bp DEL and substitutions in the CDS (Fig. 3h). This gene set encodes an NAD-dependent epimerase/dehydratase domain-containing protein.
Identification of genomic variations associated with bioclimatic variables
To identify loci potentially linked to climate adaptation, we performed genotypeâenvironment association (GEA) analysis based on SVs identified from the graph-based pangenome and small variations detected in 427 previously generated resequenced moso bamboo accessions10. We applied both latent factor mixed models 2 (LFMM2) and redundancy analysis (RDA) to identify climate-associated genetic variations using 19 bioclimatic (BIO) variables from WorldClim, including 11 temperature-related variables (BIO1âBIO11) and 8 precipitation-related variables (BIO12âBIO19) (Supplementary Data 12). We first confirmed the population structure through SNPs using ADMIXTURE and found that Kâ=â1; these results are consistent with those of previous studies10 (Supplementary Fig. 15). LFMM2 initially detected 96,638 SNPs, 7456 InDels, and 449 SVs that were significantly associated with bioclimatic variables (FDR-adjusted P-valueâ<â0.05) (Supplementary Data 13â14). Based on variable correlation and gradient forest (GF) ranking (see Methods) (Supplementary Figs. 16â17), six bioclimatic variables were selected for RDA to further filter variations. These variables included annual mean temperature (BIO1), mean diurnal range (BIO2), max temperature of warmest month (BIO5), mean temperature of wettest quarter (BIO8), precipitation of driest month (BIO14), and precipitation seasonality (BIO15). After retaining the variations identified by both the LFMM2 and RDA methods (Supplementary Fig. 18 and Supplementary Data 14â15), we identified 1050 adaptive variations (958 SNPs, 90 InDels, and 2 SVs) associated with bioclimatic variables (Supplementary Data 16), representing the core genomic variations underlying climate adaptation in moso bamboo. Additionally, compared with 123 variations related to precipitation, 996 variations were associated with temperature. RDA revealed that the contribution of climate effects explained 35% of the genetic variation in the adaptive variations, which was substantially greater than that of geography (Supplementary Table 12).
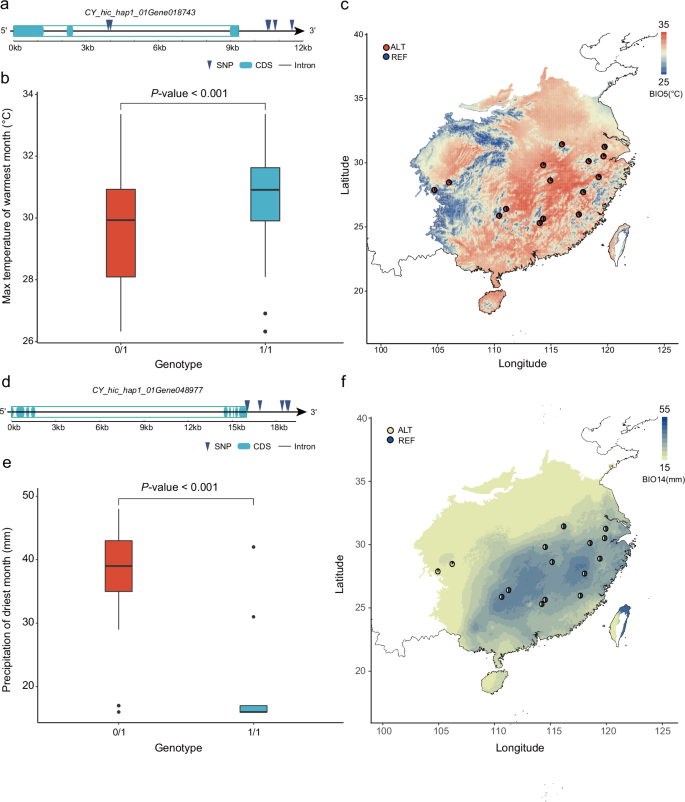
To validate climate associations and examine the potential adaptive roles of these variations, we focused on several top candidates related to temperature and precipitation. For example, we identified a SNP associated with BIO5 on chromosome 19, the most significant of which was chr19_24871064_SNP (LFMM2 FDR-adjusted P-valueâ=â0.019) (Supplementary Fig. 19). The homozygous form exhibited higher BIO5 values (Fig. 4b). In higher-BIO5 regions (e.g., XN, LY), the G allele frequency was greater, while lower-BIO5 regions (e.g., XA, HZP) showed a lower G allele frequency (Fig. 4c), with a correlation of 0.62 (Supplementary Fig. 20). This variation is located in the intron of CY_hic_hap1_01Gene018743 (Fig. 4a), and some other associated SNPs are located downstream of this gene, which encodes a heat stress-related CCHC-type zinc finger protein28. We also detected an SV (chr18_28562210_SV) on chromosome 18 associated with BIO5 (LFMM2 FDR-adjusted P-valueâ<â0.001). Accessions possessing this SV exhibited higher BIO5 values (Supplementary Fig. 21). The gene closest to this SV is CY_hic_hap1_01Gene010919, encoding the abscisic acid receptor PYL8, which plays important roles in the regulation of stress responses29.
a Locus associated with the max temperature of warmest month (BIO5) located in the intron and downstream of CY_hic_hap1_01Gene018743 on chromosome 19. d Locus associated with the precipitation of the driest month (BIO14) located downstream of CY_hic_hap1_01Gene048977 on chromosome 13. b, e Comparison of BIO5 (b) and BIO14 (e) values across different genotypes of moso bamboo accessions. The box plots show the medians (centerlines), interquartile ranges (boxes), and 1.5 times the interquartile ranges (whiskers). The BIO5 (b) nâ=â268 (0/1) and 159 (1/1), and BIO14 (e) nâ=â375 (0/1) and 50 (1/1). Statistical significance was determined using a two-sided Wilcoxon rank-sum test, with P-values of 9.29e-10â<â0.001 (b) and 1.99e-27â<â0.001 (e). c, f Allele frequencies of candidate adaptive SNPs associated with BIO5 (c: chr19_24871064_SNP) and BIO14 (e: chr13_64739621_SNP). Circles indicate allele frequencies, with red representing the alternative allele and blue representing the reference allele in (c), while yellow represents the alternative allele and blue represents the reference allele in (f). The map colors reflect variations in the BIO5 (c) and BIO14 (f) values across the distribution range of the moso bamboo population.
For precipitation-related BIO14, we identified an associated variation on chromosome 13 (Supplementary Fig. 22). The most significantly associated variation was chr13_64739621_SNP (LFMM2 FDR-adjusted P-valueâ<â0.001). The accessions carrying this variation had lower BIO14 values (Fig. 4e). In the two low-BIO14 regions of HZP and CS, the G allele frequency was greater (Fig. 4f), with a strong positive correlation (Pearsonâs râ=â0.81) (Supplementary Fig. 23). These variations are downstream of CY_hic_hap1_01Gene048977 (Fig. 4d), which is adjacent to a gene encoding a drought stress-related receptor-like protein kinase (RLK)30.
Predicting vulnerable moso bamboo populations
Using identified climate-associated variations and future climate projections, we calculated the risk of non-adaptedness (RONA), which represents the expected allele frequency offsets needed for moso bamboo to adapt to future conditions. Four general circulation models (GCMs) were considered: the Australian Community Climate and Earth System Simulator Coupled Model version 2 (ACCESS-CM2)31, the second generation CMCC Earth System Model (CMCC-ESM2)32, the Goddard Institute for Space Studies Model E version 2.1 coupled with the GISS Ocean (GISS-E2-1-G)33, and the Model for Interdisciplinary Research on Climate version 6 (MIROC6)34, which participate in the World Climate Research Programme Coupled Model Intercomparison Project Phase 6 (WCRP CMIP6) under two shared socioeconomic pathways (SSPs). The two SSPs included a low greenhouse gas emissions scenario (SSP126) and a high greenhouse gas emissions scenario (SSP585). The results revealed higher RONA values under the SSP585 high-emissions scenario than under the SSP126 low-emissions scenario. For all temperature-related variables, the RONA values were greater than those of the precipitation-related variables, and the differences between SSP585 and SSP126 were greater (Fig. 5a). For temperature, the overall trend indicated an increase in the future; therefore, we focused specifically on BIO5. Under BIO5 projections, the highest RONA occurred in XN, likely due to its current exposure to the highest temperatures and the greatest projected warming in this region (Fig. 5b). Regions LY, YF, and AJ also showed relatively high RONA values for BIO5, suggesting that protection measures against extreme heat events may be needed for these populations.
a Comparison of the mean risk of non-adaptedness (RONA, y-axis) between the SSP126 (red) and SSP585 (blue) emission scenarios from 2061â2080 across 19 bioclimatic variables (BIO1âBIO19, x-axis) and four climate models (ACCESS-CM2, CMCC-ESM2, GISS-E2-1-G, and MIROC6). The error bars represent the mean values plus or minus the standard error. b Mean RONA estimates for the moso bamboo population under the high-emission scenario (SSP585) and the max temperature of warmest month (BIO5) from 2061 to 2080 based on four individual climate models (ACCESS-CM2, CMCC-ESM2, GISS-E2-1-G, and MIROC6). The map colors indicate projected climate changes in BIO5, with darker red indicating more substantial increases in temperature. The circle size represents the RONA values of different populations. c, d Map of the gradient forest (GF)-predicted local genomic offset averaged across four climate models for the distribution of moso bamboo under SSP126 (c) and SSP585 (d) from 2061â2080. The color scale from blue to red indicates increasing genomic offset. e, f RGB map showing local (red), forward (green), and reverse (blue) genomic offsets for SSP126 (e) and SSP585 (f), respectively. Brighter cells (closer to white) have relatively greater genomic offset values, and darker cells (closer to black) have relatively lower values along each axis. The lower panels are the bivariate scattergrams of (e, f) with 1:1 lines.
Additionally, GF modeling incorporating all bioclimatic variables predicted local genomic offset across regions representing vulnerability under climate change. The greatest risks were projected in the northwestern regions (Fig. 5c, d). In addition to the local genomic offset, we also calculated the forward and reverse genomic offsets (Fig. 5e, f and Supplementary Figs. 24, 25). Figures 5e, f show a combinative visualization of three genomic offsets (local offset, forward offset, and reverse offset) by mapping them as red, green, and blue bands, respectively, in an RGB color space. Brighter cells (closer to white) and darker cells (closer to black) presented relatively greater and lower values along each axis, respectively. Most accessions from the northern regions appear brighter (Fig. 5e, f), indicating that they had relatively high offset values, suggesting that even with migration, they still face greater vulnerability compared to the southern regions. However, both the forward and reverse offsets were lower than the local offset (lower panels in Fig. 5e, f) in most of the northern region, suggesting that assisted migration may to some extent enable adaptation to future climate change. Consistent with the RONA results, all the genomic offsets were greater under the SSP585 scenario than under the SSP126 scenario, and the regions with high genomic offsets (brighter areas) were also larger, suggesting that the more extreme climate change associated with fossil fuel development (SSP585) may expose moso bamboo populations to greater adaptive challenges and potential risks than under the more sustainable scenario (SSP126). Moso bamboo plants in major natural distribution regions are still in a relatively safe position under the SSP126 scenario. However, under the SSP585 scenario, some major natural growth regions, especially the two westernmost natural growth regions, will face risks (Fig. 5f).
Discussion
The introduction and application of haplotype-resolved genomes, graph-based pangenomes, and genus-level pangenomes have greatly enriched our understanding of the genomic diversity of species or taxa, providing powerful tools for revealing the genetic basis of important traits23,35,36. In this context, our study on moso bamboo (Phyllostachys edulis), an economically and ecologically vital nontimber resource, has made significant progress by employing third-generation PacBio HiFi and Hi-C sequencing technologies to obtain the haplotype genomes of 16 representative moso bamboo accessions and construct a comprehensive pangenome. These genomic resources not only more comprehensively capture the heterogeneity of the moso bamboo genome but also provide valuable genetic information for a deeper understanding of moso bamboo adaptability to diverse environmental conditions. However, despite the significant advantages of haplotype genomes and pangenomes over traditional collapsed genomes, there are still challenges in their practical application. Mainstream omics analysis workflows, such as transcriptomics and epigenomics, still predominantly rely on aligning sequencing data to linear reference genomes, failing to fully utilize the rich diversity information contained within haplotype genomes and pangenomes. Therefore, while continuously improving the accuracy and completeness of genomic data, it is imperative to enhance the analytical framework and application environment of these high-quality genomic resources, thereby ensuring their optimal utilization.
To fully utilize these genomic resources, we integrated the haplotype genome and the pangenome, revealing valuable insights into the genomic architecture of moso bamboo. Our study revealed that the differences between the two moso bamboo haplotypes exceeded the differences between the two moso bamboo accessions. Given the asexual reproduction of moso bamboo over extended periods and its 67-year flowering cycle37,38, the primary source of variation is likely rare somatic mutations occurring within one haplotype. Asexual reproduction makes it difficult for variations accumulated in accessions to be transmitted, as the absence of meiosis prevents the exchange of genetic material between homologous chromosomes (Supplementary Fig. 26). We hypothesized that there would have been a difference between the two haplotypes in the common ancestor of moso bamboo populations in different regions and that the accumulation of somatic mutations in moso bamboo from different regions did not exceed the original difference between the two ancestral haplotypes. These factors have led to the phenomenon where, quantitatively, inter-haplotype variations exceed the genetic variations among different accessions. Additionally, we discovered that heterozygosity might be overestimated in traditional variation detection methods. When considering the haplotype genome, we found that universally heterozygous sites are also heterozygous in the reference genome and should not be regarded as variation sites between accessions. Filtering out these variations between haplotypes leads to a decrease in the detected heterozygosity, while also suggesting that genetic diversity is lower than originally estimated.
Owing to its low genetic diversity, in-depth studies on the adaptability of moso bamboo to diverse environmental conditions are important. We observed that the core-single gene sets and allele-specific expression (ASE) phenomena were closely related to environmental adaptability and identified two sets of climate-related heterozygous variation sites, which may imply that haplotypes play a significant role in the environmental adaptation of moso bamboo. Our study also showed that under the high-emissions scenario SSP585, the moso bamboo population faces significant adaptive risks (Fig. 5), highlighting the importance of emission reduction measures for alleviating the pressure of climate change. Particularly in the northwestern region, we recommend prioritizing habitat restoration where the risk is most severe (Fig. 5c) and considering assisted migration for the northern population (Fig. 5eâf) while addressing potential competition risks. Notably, our samples contained only moso bamboo from all the major natural distribution regions of moso bamboo in China, and some of the human-transplanted populations or extreme populations were not included in the study. Supplementing these populations, and even global moso bamboo accessions, could enable the identification of more variations adapted to extreme environments. For risk predictions such as RONA and local offsets that do not involve migration, the absence of these samples is less impactful. However, for forward and reverse offset analyses, incorporating additional populations could uncover regions more conducive to moso bamboo cultivation and identify moso bamboo populations better suited for migration to extreme regions. Nevertheless, the application of genomic offset in conservation planning is still in its infancy, and empirical validation of its predictions is necessary to assess its practical utility18,19. This can be achieved through carefully designed experiments, such as common garden trials or controlled environment tests, which compare genomic offset predictions with realized fitness outcomes in populations exposed to environmental change14,19.
Methods
Sample collection
To optimize the representation of genetic and environmental diversity, we selected 16 representative moso bamboo accessions (RMAs) based on a previous phylogenetic study that identified the speciesâ primary natural distribution in China10,39 (Supplementary Fig. 1). Our sampling strategy aimed to capture the extensive genetic diversity present in moso bamboo by covering all its major habitats, ensuring a comprehensive representation of the populations. The 16 RMAs, each chosen from one of the 16 regions (Supplementary Table 1 and Supplementary Fig. 1), collectively captured a wide range of variability across the moso bamboo population. Additionally, we used genetic information from 427 resequenced accessions obtained in our previous study10 (Supplementary Data 17), which covered all the main natural distribution regions of moso bamboo in China, to enhance the genetic representation of our samples in this study. In each region, moso bamboo shoots were collected for DNA extraction in April 2020. Concurrently, young leaves, stems, roots, and rhizomes were collected from the same moso bamboo rhizome as the RNA-seq samples in each region. DNA samples were rapidly dried using silica gel beads, while RNA samples were placed in an RNA stabilization solution. All samples were stored and transported on dry ice. Representative specimens were then deposited at the International Centre for Bamboo and Rattan.
Genome and transcriptome sequencing
High-molecular-weight genomic DNA (gDNA) extraction was performed with meticulous care and stringent quality control. Purity and quantity were assessed by a Nanodrop 1000 spectrophotometer and Qubit assays (Thermo Fisher Scientific, CA, USA). SMRTbell libraries with inserts of approximately 15âkb were generated using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, CA, USA) and size-selected into narrow fractions with the SageELF system (Sage Science, Beverly, MA, USA) to improve sequencing accuracy. Libraries were sequenced on 2â3 SMRT Cells 8âM on the Sequel II platform (Pacific Biosciences, CA, USA) using 30âh movie times to maximize the data yield and quality.
For chromosome-level scaffolding, Hi-C libraries were constructed according to the standard protocol and sequenced using a 150âbp paired-end strategy on the DNBSEQ-T7 platform. For RNA-seq, tissues from three biological replicates per RMA were collected, including leaves, stems, roots, and rhizomes (Supplementary Data 1). Total RNA was extracted using the cetyltrimethylammonium bromide (CTAB) protocol, followed by library construction with the NEBNext Ultra RNA Library Prep Kit and 150âbp paired-end sequencing on the DNBSEQ-T7 platform. All library preparation and sequencing for this project were performed by Annoroad Gene Technology Co., Ltd.
Genome assembly
HiFi reads were generated from subreads using the CCS algorithm v6.2.040 and assembled into contigs with Hifiasm v0.16.1-r37541. For CY, HB, and HZP, Hi-C sequencing reads were used as inputs for Hifiasm to assist in generating haplotype assemblies. Assembly quality was evaluated for structure accuracy by assessing the completeness of the Embryophyta_odb10 dataset (1614 plant orthologs) with BUSCO v5.4.342. The LTR assembly index (LAI) was calculated with LTR_retriever v2.9.043 and a mutation rate of 8.51âÃâ10â8â38, with an estimated moso bamboo genome size of 1.92âGb10. Meryl databases were generated with Meryl v1.4144 for the raw sequencing reads and the assemblies. The quality value (QV) and k-mer completeness were then calculated using Merqury v1.344 by comparing the k-mer spectra of the assembly and the raw reads. Hi-C reads were used to anchor scaffolds to chromosomes via the 3D-DNA pipeline v17012345. The resulting contact maps were visualized with Juicerbox v1.5.246.
Genome annotation
Transposable element (TE) annotation combines de novo and homology-based approaches at the DNA and protein levels. A de novo repeat library was constructed with RepeatModeler v2.0.347, and RepeatMasker v4.1.2-p148 was used to classify TEs by mapping to the library and Repbase v21.1249. For gene prediction, a combination of homology-based, RNA-seq-based, and ab initio methods was utilized. Homologous proteins from seven plant species (Ananas comosus, Arabidopsis thaliana, Glycine max, Hordeum vulgare, Nicotiana attenuata, Oryza sativa, and Zea mays) were downloaded from Phytozome v1150 and aligned to the genome using GeneWise v2.4.151. The RNA-seq reads were preprocessed by Trimmomatic v0.3852 and mapped to the genome using HISAT2 v2.1.053. AUGUSTUS v3.4.054 was used to perform ab initio predictions trained on the transcriptome. GETA v2.5.5 (https://github.com/chenlianfu/geta) was used to integrate evidence from all methods to generate a high-quality gene set for each accession. BUSCO v5.4.3 was used to assess gene set completeness. Functional annotation was assigned to the genes using DIAMOND55 according to the best hits to the following databases: NCBI Nr v2021082456, Swiss-Prot v2021082457, KOG v2003072158, eggNOG v5.059, InterPro v8560, Pfam-A v34.061, GO (integrated with eggNOG and InterPro), and KEGG databases62 (based on KAAS v2.163). All databases were accessed on 11 December 2021.
Switch error calculation
Switch errors were calculated using calc_switchErr v1.021. Briefly, PacBio HiFi reads were aligned to haplotype 1 using pbmm2 v1.5.0 (https://github.com/PacificBiosciences/pbmm2), and SNPs were identified using DeepVariant v1.4.064. SNP phasing was subsequently performed on Whatshap v1.165, followed by alignment of the two haplotype genomes using nucmer in MUMmer4 v4.0.0rc166. SNPs were identified using the show-snps command, and switch errors were calculated by comparing the SNPs to those identified in the aligned reads. For accessions with Hi-C data (CY, HB, and HZP), Hi-C sequencing reads were aligned to haplotype 1 using bwa v0.7.1767, SNPs were called using DeepVariant, and switch errors were calculated from both the HiFi and Hi-C SNPs.
Allele identification
The determination of alleles between any two haplotype assemblies was based on a combination of protein sequence similarity and relative position. Protein sequence similarity was calculated by aligning every haplotype genome using BLASTP v2.9.0â+â68. The relative positions between two genes from different genomes were determined by aligning the assemblies using Minimap2 v2.24-r112269, and gene pairs within 40âkb (the average gene distance in the moso bamboo genome) were retained for further analysis. The 40âkb threshold was chosen because it approximates the average gene distance in the moso bamboo genome, which has a size of approximately 2âGb and contains approximately 50,000 genes. This threshold allows for some positional flexibility, permitting alleles to be separated by up to one average gene interval. Based on the criteria of closer genomic distance and higher sequence identity, the best reciprocal alignments were preserved as allele pairs. Genes that met the similarity threshold for the other haplotype but were not among the best-aligned allele pairs were identified as haplotype-specific duplicated genes (Supplementary Data 18). To promote transparency and reproducibility, the script for this step has been made publicly available on GitHub (https://github.com/ZhaoGroupLab/moso-bamboo-pangenome).
Pangenome construction
A pangenome was constructed by iteratively incorporating genes from each genome into aggregated gene sets. Any genome could serve as the initial reference. For each additional genome, its genes were mapped to allele sets in the existing pangenome based on identified alleles. Genes without a mapped allele were placed into new sets. This process was repeated until all the genomes were incorporated. Pangenome aggregation curves were generated by permuting the order of genome addition.
Definitions of gene sets in pangenomes
Pangenome gene sets were categorized based on their frequency across accessions. The core gene set contained genes present in all 16 accessions. The softcore gene set included genes present in 12â15 accessions. The dispensable gene set contained genes present in 2â11 accessions. The private gene set included genes present in only a single accession. Additionally, to characterize the presence and absence of alleles in the haplotypes, we classified all the gene sets into three types based on the alleles. If every gene in a gene set had its allele present, we defined this as a double-allele set. In contrast, if none of the genes in a set had alleles present, we defined this as a single-allele set. The third type fell between the double-allele and single-allele sets. If some genes in a set had alleles present while others did not, we defined this as a variable-allele set. In addition, single-allele gene sets containing duplicated genes were reclassified as variable-allele gene sets. Therefore, considering both accession occurrence and allele presence, we ultimately divided all the gene sets into 12 categories: core-double, core-variable, core-single, softcore-double, softcore-variable, softcore-single, dispensable-double, dispensable-variable, dispensable-single, private-double, private-variable and private-single.
Allele-specific expression
Allele-specific expression (ASE) analysis was performed on triplicate RNA-seq data from 3 or 4 tissues (leaves, stems, rhizomes, and roots) collected from 16 representative moso bamboo accessions. Reads were preprocessed with Trimmomatic v0.39 and aligned to the corresponding haplotype assemblies using HISAT2 v2.1.0. The transcripts per kilobase million (TPM) values were calculated with StringTie v1.3.570, and the alleles were compared using DESeq2 v1.34.071. Alleles with a P-valueâ<â0.05 and an absolute log2(fold change) > 1 were classified as having ASE.
SNP and InDel identification
We identified SNPs and InDels based on whole-genome alignment and read alignment strategies. For whole-genome alignment strategies, haplotype assemblies were aligned to the CY haplotype 1 reference using nucmer in MUMmer v4.0.0rc1. SNPs were identified using the show-snps command, and InDels were identified using svim-asm v1.0.272. For the read alignment strategies, PacBio HiFi reads were aligned to haplotype 1 using pbmm2 v1.5.0 (https://github.com/PacificBiosciences/pbmm2), and SNPs and InDels were identified using DeepVariant v1.4.063. Subsequently, the SNPs and InDels detected by both strategies were retained. Variations were converted to reference genome coordinates based on query-reference comparisons. The BCFtools v1.973 merged command was used to integrate SNPs and InDels across accessions.
Structural variation identification
Genome-wide structural variations (SVs) were identified using five pipelines based on read alignment or whole-genome alignment to the CY haplotype 1 reference: (i) Minimap2 v2.24-r1122 + cuteSV v1.0.1374; (ii) Ngmlr v0.2.775 + sniffles v1.0.1275; (iii) pbmm2 v1.5.0 + pbsv v2.8.0 (https://github.com/PacificBiosciences/pbsv); (iv) Minimap2 + svim-asm v1.0.2; and (v) Nucmer v4.0.0rc166 + Assemblytics v1.2.176.
Filtering and merging of SVs
SVs were filtered and merged within and across accessions based on the above five pipelines. Initially, a given SV required support from at least two callers to be retained. SVs >50âkb were specifically detected by at least one genome-wide alignment method. After filtering, SVs were merged as follows: deletions (DELs) and inversions (INVs) within 1% of the length of the larger SV (with a minimum of 5âbp and a maximum of 100âbp) were merged. Insertions (INSs) within 10âbp of each other with >â90% BLAST alignment identity and coverage were merged.
Identification of the frequency of variations in genomic regions
The frequencies of SVs and short variations (SNPs and InDels) were identified by dividing the genome into 100-kb genomic windows using the makewindows utility in BEDtools v2-2.25.077. The coverage command was used to calculate the number of mutations per window. The windows were sorted by mutation count in descending order.
Graph-based pangenome construction
A graph-based pangenome was constructed with vg v1.3878 through the following steps. The vg construct command was used to construct the initial graph. The gbwt command was used to generate the.gbwt file. The snarls command was used to produce the.snarls file. The index command built distance index files and minimized the index. Additionally, short reads from 427 resequenced accessions were aligned to the graph-based pangenome using vg giraffe79.
SNP and InDel calling based on resequenced reads
The raw sequencing reads were processed using the same pipeline as in our previous study to ensure consistency. Briefly, the filtered resequenced reads were aligned to the CY haplotype 1 reference genome using BWA v0.7.17. Aligned reads (BAM files) were sorted using SAMtools v1.980, and duplicates were removed using GATK v4.2.081. SNP and InDel calling was performed using the joint calling method within GATK. We obtained the genomic variant call format (GVCF) in ERC mode for each accession based on reads. Then, we filtered SNPs directly based on quality, removing variations with a quality score lower than 50 based on the quality score distribution.
To determine a pruned SNP set, we used PLINK v1.982. The resulting SNPs were then used to assess population structure using ADMIXTURE v1.3.083 for multiple repeats with different random seeds. The population structure analysis showed Kâ=â1 (Supplementary Fig. 15), which was consistent with our previous findings10.
Identification of climate-associated variations
The SVs identified from the graph-based pangenome were constructed using 16 representative moso bamboo accessions, and the SNPs and InDels detected from the previously generated 427 resequenced moso bamboo accessions were used for the identification of climate-associated variations. We retained variations with a minor allele frequency greater than 0.05, a maximum missing data proportion of 0.2, and a minimum depth of 3 across all variation types, including SNPs, InDels, and SVs, using VCFtools v0.1.1384. Since most variations occurred between haplotypes rather than within accessions in the moso bamboo population, we filtered sites with minor genotype frequencies <â0.05, leaving 1,467,461 SNPs, 103,955 InDels and 4,643 SVs for genome-wide identification of climate-associated variations. We tested for correlations with 19 bioclimatic variables (BIOs) using latent factor mixed models 2 (LFMM2) implemented in the R package LEA v3.6.085. Based on the ADMIXTURE v1.3.0 results and the results of a previous study10, we set Kâ=â1 for LFMM2 analyses and used false discovery rate (FDR) correction for multiple testing to obtain adjusted P-values. We retained genetic variations with an adjusted P-valueâ<â0.05 as climate-associated variations. Additionally, we performed gradient forest (GF) analysis in the R package gradientForest v0.1.3486 to rank the importance of the 19 BIOs and examined the correlations among them. Six BIOs with correlation coefficients |râ|â<0.6 (BIO1 (representing BIO1, BIO6, BIO9 and BIO11), BIO2 (representing BIO2), BIO5 (representing BIO5 and BIO10), BIO8 (representing BIO8), BIO14 (representing BIO12, BIO13, BIO14, BIO16, BIO17, BIO18 and BIO19) and BIO15 (representing BIO15)) were selected for redundancy analysis (RDA) using the R package vegan v2.6.487. Significant variations were defined as loadings exceeding 3.5 standard deviations from the mean along one or more RDA axes. The climate-associated adaptive variations were identified by retaining the variations detected by both methods, LFMM2 and RDA, for each climatic variable. In addition, we used RDA and partial RDA (pRDA) to quantify the relative contributions of geography and the environment. The longitude and latitude values characterized the explanatory variables of geography, and the 6 BIOs used in the above RDA characterized the explanatory variables of climate.
Calculation of RONA and genomic offset
Future climate projections were obtained using four general circulation models (GCMs) from the World Climate Research Programme Coupled Model Intercomparison Project Phase 6 (WCRP CMIP6): the Australian Community Climate and Earth System Simulator Coupled Model version 2 (ACCESS-CM2), second-generation CMCC Earth System Model version 2 (CMCC-ESM2), the Goddard Institute for Space Studies Model E version 2.1 coupled with the GISS Ocean (GISS-E2-1-G), and the Model for Interdisciplinary Research on Climate version 6 (MIROC6). Two shared socioeconomic pathways (SSPs) were consideredâa low-emission scenario (SSP126) and a high-emission scenario (SSP585)âfor two 20-year periods (2061â2080 for SSP126 and 2081â2100 for SSP585). The SSPs represent combinations of shared socioeconomic pathways and representative concentration pathways (RCPs). SSP126 is an abbreviation for the SSP1-RCP2.6 scenario. SSP1 (sustainability, taking the green road) assumes a gradual shift toward a more sustainable world, with emphasis on human well-being and reduced inequality. RCP2.6 represents one mitigation scenario leading to a very low forcing level88,89. Similarly, SSP585 is the shortest form for the SSP5-RCP8.5 scenario. SSP5 (fossil fuel development, taking the highway) assumes rapid economic growth driven by fossil fuels, with high energy demand and limited efforts to mitigate greenhouse gas emissions. RCP8.5 represents a very high-baseline emission scenario88,89.
The relative risk of non-adaptedness (RONA) was calculated for all the sampling regions under the SSP126 and SSP585 scenarios using pyRona v0.3690. Gradient forest (GF) analysis was then performed in R to predict genomic offset under the different future scenarios using 19 BIOs. We Subsequently calculated the local, forward, and reverse offsets according to the methodology13, the script for this step has been made publicly available on GitHub (https://github.com/ZhaoGroupLab/moso-bamboo-pangenome). Local offset, a measure of the vulnerability of a resident population to climate change, was calculated by estimating the predicted change in allele frequencies at climate-adaptive loci that was necessary for the population to adapt to local climate changes over time. In contrast, forward offset assumed that populations had unlimited migration ability. It was calculated by identifying the minimum predicted offset if propagules or alleles could move, through gene flow, to any suitable habitat within the range. Reverse offset represented the possibility that any population in the current range would be preadapted to a particular location in the future. Reverse offset was calculated by identifying the minimum offset between hypothetical populations within the current range in the future climate and populations in the current climate. For the forward and reverse offsets, we set the migration distance to infinity because moso bamboo propagates vegetatively for a long period and is primarily introduced through transplanting.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
For the 427 resequenced accessions from our previous study, the sequencing data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under accession number PRJNA755164 and the China National GenBank (CNGB) under accession number CNP0001535. For the 16 representative moso bamboo accessions and 186 RNA-seq samples generated in this study, the sequencing data have been deposited in the Genome Sequence Archive (GSA) under accession numbers CRA014344. The reference genome used in this study is available in the GSA database under BioProject code PRJCA022610. The source data underlying Figs. 1aâd, 2aâf, 3aâe, 3g, 4bâc, 4eâf, 5aâf, and the Supplementary Figs. are provided as source data files. Source data are provided with this paper.
Code availability
All software used in our study are publicly available, and the specific parameters are provided in Supplementary Table 13. The customized codes used in this study have been deposited in GitHub [https://github.com/ZhaoGroupLab/moso-bamboo-pangenome] and Zenodo [https://doi.org/10.5281/zenodo.12794412]91.
References
Ramakrishnan, M. et al. Genetics and genomics of moso bamboo (Phyllostachys edulis): current status, future challenges, and biotechnological opportunities toward a sustainable bamboo industry. Food Energy Secur. 9, e229 (2020).
China Forestry and Grassland Administration. Development Plan for Forestry and Grassland Industry (2021â2025). (2019).
China Forestry and Grassland Administration et al. Opinions from Ten Departments on Accelerating the Innovative Development of the Bamboo Industry. (2021).
Zhou, G., Meng, C., Jiang, P. & Xu, Q. Review of carbon fixation in bamboo forests in China. Bot. Rev. 77, 262â270 (2011).
Frankham, R., Briscoe, D. A. & Ballou, J. D. Introduction to conservation genetics. (Cambridge university press, 2002).
Willi, Y., Buskirk, J. V. & Hoffmann, A. A. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 37, 433â458 (2006).
Warren, R. et al. Quantifying the benefit of early climate change mitigation in avoiding biodiversity loss. Nat. Clim. Change 3, 678â682 (2013).
Alcala, N., Streit, D., Goudet, J. & Vuilleumier, S. Peak and persistent excess of genetic diversity following an abrupt migration increase. Genetics 193, 953â971 (2013).
Aitken, S. N., Yeaman, S., Holliday, J. A., Wang, T. & Curtis-McLane, S. Adaptation, migration or extirpation: climate change outcomes for tree populations. Evol. Appl. 1, 95â111 (2008).
Zhao, H. et al. Analysis of 427 genomes reveals moso bamboo population structure and genetic basis of property traits. Nat. Commun. 12, 5466 (2021).
Yang, S., Zhang, Y., Sun, M., Goldstein, G. & Cao, K. Recovery of diurnal depression of leaf hydraulic conductance in a subtropical woody bamboo species: embolism refilling by nocturnal root pressure. Tree Physiol. 32, 414â422 (2012).
Arend, M. et al. Lack of hydraulic recovery as a cause of post-drought foliage reduction and canopy decline in European beech. N. Phytol. 234, 1195â1205 (2022).
Gougherty, A. V., Keller, S. R. & Fitzpatrick, M. C. Maladaptation, migration and extirpation fuel climate change risk in a forest tree species. Nat. Clim. Change 11, 166â171 (2021).
Sang, Y. et al. Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia. Nat. Commun. 13, 6541 (2022).
Hung, T. H. et al. Range-wide differential adaptation and genomic offset in critically endangered Asian rosewoods. Proc. Natl Acad. Sci. USA 120, e2301603120 (2023).
Fitzpatrick, M. C. & Keller, S. R. Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol. Lett. 18, 1â16 (2015).
Chen, Y. et al. The combination of genomic offset and niche modelling provides insights into climate change-driven vulnerability. Nat. Commun. 13, 4821 (2022).
Gain, C. et al. A quantitative theory for genomic offset statistics. Mol. Biol. Evol. 40, msad140 (2023).
Lotterhos, K. E. Interpretation issues with âgenomic vulnerabilityâ arise from conceptual issues in local adaptation and maladaptation. Evol. Lett. 8, 331â339 (2024).
Neale, D. B. & Kremer, A. Forest tree genomics: growing resources and applications. Nat. Rev. Genet. 12, 111â122 (2011).
Zhang, X. et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 53, 1250â1259 (2021).
Sun, Y., Shang, L., Zhu, Q.-H., Fan, L. & Guo, L. Twenty years of plant genome sequencing: achievements and challenges. Trends Plant Sci. 27, 391â401 (2022).
Shi, T. et al. The super-pangenome of Populus unveils genomic facets for its adaptation and diversification in widespread forest trees. Mol. Plant 17, 725â746 (2024).
Yu, J. et al. Analysis of aldoâketo reductase gene family and their responses to salt, drought, and abscisic acid stresses in Medicago truncatula. Int. J. Mol. Sci. 21, 754 (2020).
Boyes, D. C., Nam, J. & Dangl, J. L. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl Acad. Sci. USA 95, 15849â15854 (1998).
Zhang, M., Wang, L. & Zhong, D. Photolyase: dynamics and mechanisms of repair of sun-induced DNA damage. Photochem. Photobiol. 93, 78â92 (2017).
Mishina, T. E. & Zeier, J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141, 1666â1675 (2006).
Sun, A. et al. Comprehensive genome-wide identification, characterization, and expression analysis of CCHC-type zinc finger gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 13, 892105 (2022).
Di, F. et al. Genome-wide analysis of the PYL gene family and identification of PYL genes that respond to abiotic stress in Brassica napus. Genes 9, 156 (2018).
Chen, X. et al. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 63, 53â78 (2021).
Bi, D. et al. Configuration and spin-up of ACCESS-CM2, the new generation Australian Community Climate and Earth System Simulator Coupled Model. J. South. Hemisph. Earth Syst. Sci. 70, 225â251 (2020).
Lovato, T. et al. CMIP6 simulations with the CMCC Earth System Model (CMCC-ESM2). J. Adv. Model. Earth Syst. 14, e2021MS002814 (2022).
Kelley, M. et al. GISS-E2.1: configurations and climatology. J. Adv. Model. Earth Syst. 12, e2019MS002025 (2020).
Kataoka, T. et al. Seasonal to decadal predictions with MIROC6: description and basic evaluation. J. Adv. Model. Earth Syst. 12, e2019MS002035 (2020).
Sun, H. et al. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nat. Genet. 54, 342â348 (2022).
Qin, P. et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell 184, 3542â3558 (2021).
Isagi, Y. et al. Clonal structure and flowering traits of a bamboo [Phyllostachys pubescens (Mazel) Ohwi] stand grown from a simultaneous flowering as revealed by AFLP analysis. Mol. Ecol. 13, 2017â2021 (2004).
Ma, P. et al. Negative correlation between rates of molecular evolution and flowering cycles in temperate woody bamboos revealed by plastid phylogenomics. BMC Plant Biol. 17, 260 (2017).
Jiang, W. et al. Microsatellite markers revealed moderate genetic diversity and population differentiation of moso bamboo (Phyllostachys edulis)âa primarily asexual reproduction species in China. Tree Genet. Genomes 13, 130 (2017).
Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155â1162 (2019).
Cheng, H. et al. Haplotype-resolved assembly of diploid genomes without parental data. Nat. Biotechnol. 40, 1332â1335 (2022).
Manni, M., Berkeley, M. R., Seppey, M. & Zdobnov, E. M. BUSCO: assessing genomic data quality and beyond. Curr. Protoc. 1, e323 (2021).
Ou, S., Chen, J. & Jiang, N. Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 46, e126 (2018).
Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 21, 245 (2020).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92â95 (2017).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95â98 (2016).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl Acad. Sci. USA 117, 9451â9457 (2020).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinforma. 25, 4.10.11â14.10.14 (2009).
Bao, W., Kojima, K. K. & Kohany, O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178âD1186 (2012).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907â915 (2019).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114â2120 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357â360 (2015).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637â644 (2008).
Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366â368 (2021).
OâLeary, N. A. et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733âD745 (2016).
The UniProt Consortium. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 51, D523âD531 (2023).
Koonin, E. V. et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5, R7 (2004).
Hernández-Plaza, A. et al. eggNOG 6.0: enabling comparative genomics across 12 535 organisms. Nucleic Acids Res. 51, D389âD394 (2023).
Blum, M. et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 49, D344âD354 (2021).
Mistry, J. et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412âD419 (2020).
Kanehisa, M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480âD484 (2008).
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C. & Kanehisa, M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182âW185 (2007).
Poplin, R. et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983â987 (2018).
Martin, M. et al. WhatsHap: fast and accurate read-based phasing. Preprint at bioRxiv https://doi.org/10.1101/085050 (2016).
Marçais, G. et al. MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol. 14, e1005944 (2018).
Li, H. & Durbin, R. Fast and accurate long-read alignment with BurrowsâWheeler transform. Bioinformatics 26, 589â595 (2010).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinforma. 10, 421 (2009).
Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics 37, 4572â4574 (2021).
Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Heller, D. & Vingron, M. SVIM-asm: structural variant detection from haploid and diploid genome assemblies. Bioinformatics 36, 5519â5521 (2021).
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987â2993 (2011).
Jiang, T. et al. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 21, 189 (2020).
Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461â468 (2018).
Nattestad, M. & Schatz, M. C. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics 32, 3021â3023 (2016).
Borromeo, M. D. et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 16, 1259â1272 (2016).
Garrison, E. et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat. Biotechnol. 36, 875â879 (2018).
Sirén, J. et al. Pangenomics enables genotyping of known structural variants in 5202 diverse genomes. Science 374, abg8871 (2021).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491â498 (2011).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559â575 (2007).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19, 1655â1664 (2009).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156â2158 (2011).
Frichot, E. & François, O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 6, 925â929 (2015).
Ellis, N., Smith, S. J. & Pitcher, C. R. Gradient forests: calculating importance gradients on physical predictors. Ecology 93, 156â168 (2012).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.6.4. (2022).
OâNeill, B. C. et al. The Scenario Model Intercomparison Project (ScenarioMIP) for CMIP6. Geosci. Model Dev. 9, 3461â3482 (2016).
Riahi, K. et al. The Shared Socioeconomic Pathways and their energy, land use, and greenhouse gas emissions implications: an overview. Glob. Environ. Change 42, 153â168 (2017).
Pina-Martins, F., Baptista, J., Pappas, G. J. & Paulo, O. S. New insights into adaptation and population structure of cork oak using genotyping by sequencing. Glob. Change Biol. 25, 337â350 (2019).
Hou, Y. et al. Haplotype-based pangenomes reveal genetic variations and climate adaptations in moso bamboo populations. Zenodo https://doi.org/10.5281/zenodo.12794412 (2024).
Acknowledgements
This work received financial support from the National Key Research and Development Program of China (2021YFD2201000 to H.Z.), the National Natural Science Foundation of China (32271975 and 31971733 to H.Z.), the High-End Foreign Expert Recruitment Program of the PRC Ministry of Science and Technology (G2022052002L to H.Z.), and the Research and Demonstration of Key Technologies for âBamboo as Substitutes for Plasticâ in Pilot Member States of the International Bamboo and Rattan Organization (to H.Z.).
Author information
Authors and Affiliations
Contributions
Y.H. led the main bioinformatics and statistical analyses of the data and wrote the manuscript. J.G. contributed to the pangenome analyses and figures. Z.F. contributed to the genomic offset analyses. L.S. and Y.W. contributed to the genome assembly and sample collection. S.L., P.B., and B.C. contributed to the expression data analyses. V.G. and R. K. V. provided valuable insights and suggestions for the project. H.Z. conceived and supervised the entire project, provided direction and guidance, and revised the manuscript. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Octavio Paulo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisherâs note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the articleâs Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the articleâs Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hou, Y., Gan, J., Fan, Z. et al. Haplotype-based pangenomes reveal genetic variations and climate adaptations in moso bamboo populations. Nat Commun 15, 8085 (2024). https://doi.org/10.1038/s41467-024-52376-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52376-5