Abstract
A common approach to monitoring the status of physical and biological systems is through the regular measurement of various system parameters. Changes in a systemâs underlying dynamics manifest as changes in the behaviour of the observed time series. For example, the transition from healthy cardiac activity to ventricular fibrillation results in erratic dynamics in measured electrocardiogram (ECG) signals. Identifying these transitionsâchange point detectionâcan be valuable in preparing responses to mitigate the effects of undesirable system changes. Here, we present a data-driven method of detecting change points using a phase space approach. Delay embedded trajectories are used to construct an âattractor networkâ, a discrete Markov-chain representation of the systemâs attractor. Once constructed, the attractor network is used to assess the level of surprise of future observations where unusual movements in phase space are assigned high surprise scores. Persistent high surprise scores indicate deviations from the attractor and are used to infer change points. Using our approach, we find that the attractor network is effective in automatically detecting the onset of ventricular fibrillation (VF) from observed ECG data. We also test the flexibility of our method on artificial data sets and demonstrate its ability to distinguish between normal and surrogate time series.
Similar content being viewed by others
Introduction
The analysis, prediction and detection of change points in dynamical systems is an open problem that has been studied extensively in many diverse contexts such as financial systems1,2, climate dynamics3,4, ecological systems5 and medicine6,7. The existence of these change points is ubiquitous in many naturally occurring systems and describes points in time during which the behaviour of the system is altered. This difference in behaviour may be attributed to endogenous or exogenous perturbations to the system. In some cases, such change points signal the transition of the system into a different, potentially undesirable regime of dynamics where the change point is also described as a âtipping pointâ in some fields.
A related problem within the context of dynamical systems is the automatic prediction or detection of imminent change points from observed time series data. This is particularly useful when an analytical mathematical model of the system is not readily available. In medicine, dynamical systems methods are used for the automated detection of acute pathological problems such as arrhythmia and ventricular fibrillation from ECG signals8, or identifying the onset of epileptic seizures in EEG analysis9,10. Change detection is also present in non-biological problems such as predicting transitions in paleo-climatic data11,12,13, and identifying equipment failure14. The generality and universality of this problem have been long recognised and have given rise to the broader term of âchange point detectionâ to describe this commonly occurring challenge.
Change point detection is a class of problems within the domain of time series analysis primarily concerned with the detection of changes in the dynamics of an underlying system15,16,17. Typically, the aim is to accurately detect changes in a system by analysing an incoming stream of observed time series15. Effective change point detection methods are useful in automating processes for faster cursory analyses. For systems where the state of components varies over time, employing a dynamical approach to detect subtle changes in the underlying dynamics may even provide a method to preemptively diagnose the occurrence of system failure.
Change point detection methods can be classified according to two features: (1) offline vs. online, and (2) supervised vs. unsupervised methods. The first classification concerns the time period during which the detection algorithm is applied. Offline detection methods focus on the analysis of observed time series after the change has occurred (i.e., post-system failure), likened to a post-mortem of observed time series15 and are useful for labelling tasks and time-insensitive applications. In contrast, online methods analyse incoming streams of observations in real-time and flag changes as they occur. The latter method is most used for automated detection.
The second classification on supervision describes whether ground truth labels are known a priori. Supervised methods construct a reference model based on a pre-defined ground truth (e.g., user-provided labels of normal vs. abnormal)15. Here, we use the term âmodelâ loosely to describe any constructed system, set of statistics or parameters that characterise the ground truth state. Deviations from the model are then used to infer system changes.
In contrast, unsupervised methods attempt to circumvent the requirement of a pre-defined ground truth by comparing new incoming observations against recently observed data. This is usually achieved by comparing statistics (mean, probability density, permutation entropy) of data between two moving windows (subsequences) in the time series separated by some lag. A change point is flagged if the statistics of the most recent observed window greatly differ from prior observations. Some methods used for change point detection include decision trees18, support vector machines19,20,21, and statistical approaches such as Gaussian mixed models22, Gaussian processes23 and Bayesian methods24. However, the usage of statistical measures typically relies on the data adhering to some stationary distribution. This does not account for temporal dependencies often present in dynamical systems, which may be useful in uncovering and characterising changes in the underlying data generating process25. For the scope of this paper, we will focus specifically on an online supervised change point detection method.
An alternative method of identifying change points is to employ a phase space approach to analysing observed time series26,27,28,29. The temporal behaviour of a given system can be represented as a trajectory moving through phase space where axes are defined as the observed system variables. When operating under the normal regime, the trajectory of a system may settle and move along a well-defined region of space, termed an âattractorâ. Changes in the underlying system will inevitably result in changes in the dynamics of the observed time series and the resulting trajectory25,28,29. Consequently, this may cause trajectories to drift and settle into a different attractor corresponding to the new system dynamics. Therefore, the problem of change point detection may be re-framed as detecting changes in the attractor of the system. This approach evaluates and measures phase space dissimilarities between two windows of time series and draws similarities to the kernel change detection (KCD) algorithm19 where changes are flagged when the metric distance between two descriptors of the data exceeds a threshold. Several algorithms based on dynamical systems tools such as cross-recurrence plots30 and empirical mode decomposition31 have also been implemented by da Costa et al. into the Massive Online Analysis (MOA) software package25.
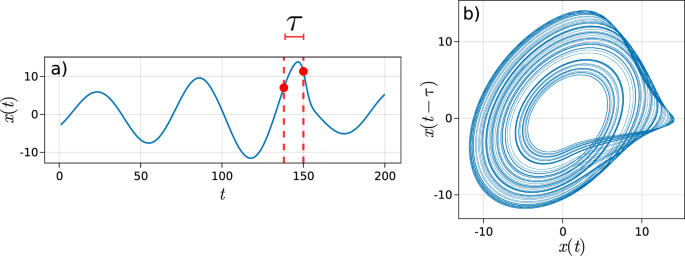
In order to employ a phase space approach, the phase space must first be reconstructed from the observed time series. This can be achieved via embedding methods such as time delay embedding. Takensâ theorem and its sequels provide basic dynamical guarantees for extracting the phase space dynamics of a deterministic system from scalar univariate time series using an appropriate time delay embedding32 (see Fig. 1 and âMethodsâ). We note that in most cases, signals from a physical system may be noisy or be driven by stochastic forces. However, time delay embedding should still provide a reasonable approximation of the dominant phase space dynamics of the system.
a Two points from the time series separated with a delay lag of Ï used to construct the delay vector. b The resulting reconstructed phase space of the system from plotting the trajectory in lagged coordinates (x(Ï),âx(tâââÏ)). (see Supplementary Note 2 for more information).
Whilst the transformation from time series to reconstructed phase space is relatively straightforward (subject to appropriately selected delay lags), achieving a faithful and parsimonious representation of the attractorâs structure and dynamics within phase space (reconstructed or otherwise) is challenging as these attractors are defined continuously in phase space. St Luce and Sayama33 proposed a method of achieving a transition network representation of an attractor by discretising phase space into voxels. This greatly simplifies the task of dealing with discrete observations of continuous data. However, such an approach does not scale well with higher dimensional data as the number of voxels increases rapidly. This problem becomes apparent when attempting to extend their approach to analysing high dimensional data such as those resulting from a high dimensional time delay embedding.
Several methods have been proposed to transform time series (and by extension dynamical attractors) into a network representation with some examples including cycle networks34, recurrence networks1,35, ordinal partition networks36,37, k-nearest neighbour network38 and visibility graphs39. Provided there is sufficient data to estimate state transition probabilities, many of these transformations extend well to higher dimensional data. But they preserve differing amounts of the spatial and dynamical information contained within the time series, both of which can be crucial in detecting subtle changes in the systemâs dynamics.
The use of a geometrical approach has also inspired the application of various topological data analysis (TDA) methods for system characterisation and change point detection. Persistent homology has previously been found to be useful in characterising the state of dynamical systems40, and also experimentally for classifying breathing signals41 and chatter detection42. More recently, persistent homology methods have also been adapted for the purpose of change point detection from temporal data. For a sequence of point clouds, mappings of Betti sequences43 and Wasserstein distances between consecutive persistence diagrams44 have been used as measures for identifying change points. Building on this, persistence diagram-based change-point detection (PERCEPT) utilising â2 divergences between persistence histograms45,46 was proposed as an improvement on previous methods and was used to analyse solar flare images. However, whilst sequences of points clouds can be constructed from scalar time series data (e.g., via sliding windows), systems whose attractors have highly complex geometry may require large windows in order for the underlying geometry to be well captured by persistent homology. This consideration in conjunction with the the poor computational scaling of persistent homology algorithms, and the quick detection response time typically desired in online change point detection tasks can make direct application of these methods challenging.
We present in this paper a method for constructing a parsimonious network representation of attractors in phase space, termed the âattractor network", that encodes both the spatial and dynamical information of the observed time series. The attractor network consists of two components: (1) the spatial network, which encodes positional information in phase space, and (2) the dynamics network, which encodes the transition frequencies and probabilities between points in state space. Functionally, the attractor network behaves as a Markov chain surrogate model of the underlying system where the adjacency matrix of the dynamics network is the transition matrix. However, attractor networks build upon the Markov chain idea by preserving the spatial positions of each node. We use the term surrogate in this case as predictions from the Markov model will always be stochastic, even if the input data is deterministic.
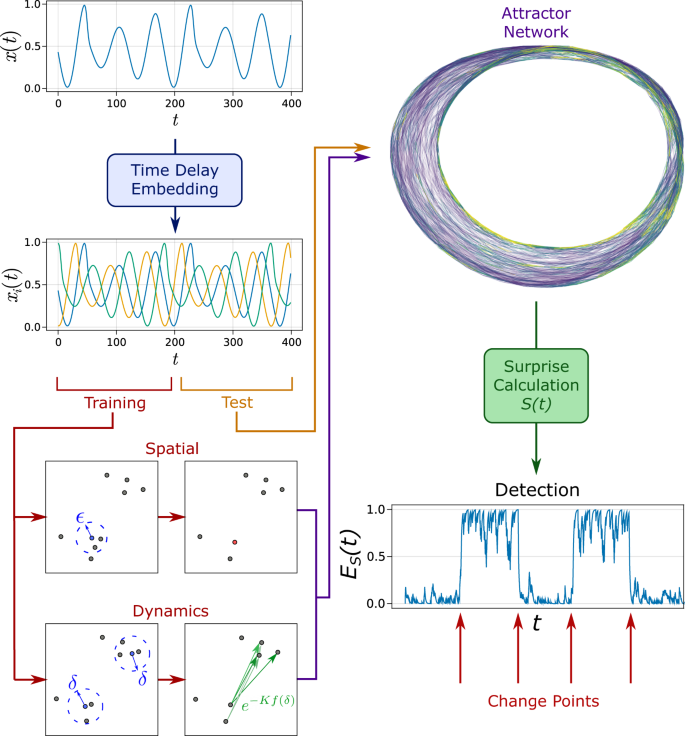
Our proposed attractor network approach is shown to be effective in tackling the challenges of the phase space approach for change point detection. Namely, our method remains parsimonious with increasing dimension whilst maintaining the network approach of discretising continuous phase space. To achieve this, attractor networks that approximately model the observed systemâs dynamics are constructed (equivalent to parametric model training) using observed data of a given system that is known to be operating within its normal regime. The resulting attractor network is subsequently used to test future incoming data observations for change points/abnormal behaviour in real time. This is done by comparing incoming observed transitions (e.g., \(\overrightarrow{x}(t)\to \overrightarrow{x}(t+\delta t)\)) against the expected transition probabilities of the system in the normal state. A measure of the surprise S(t) of each observed transition can be calculated using information theory approaches (see âMethodsâ). Unexpected transitions such as those that are infrequently or never previously observed when constructing the attractor are given high surprise values (see Fig. 2). A critical threshold S* taken as the 95% quantile of S(t) from the training data is used to convert the continuous measure of S(t) into a binary time series \(\hat{S}(t)\). We define change points as the occurrence of successive high suprise values. To detect change points, exponential smoothing is first applied to \(\hat{S}(t)\) to calculate a normalised measure of surprise ES(t). Change points then correspond to times where E(t)â>âE* where E* is a threshold defined from E(t) of the training data. A schematic overview of the analysis method is provided in Fig. 3.
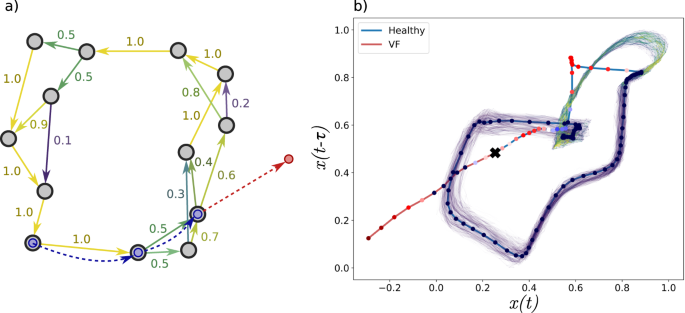
a Illustration of a constructed attractor network with edges weighted by approximate transition probabilities. Observed states are given by four red and blue nodes with the resulting trajectories shown as dashed line. Node observations are coloured according to surprise value S(t) (red is high surprise). Trajectories (dashed) are coloured either red or blue based on the classification of normal or unhealthy. Red observed states and trajectories (i.e., high S(t) and ES(t)) describe unexpected trajectories that enter into a previously unvisited part of phase space. Persistent high surprise values are indicative of change points. b Attractor network constructed from a delay embedded ECG with a trajectory showing the transition from normal to the ventricular fibrillation (VF) regime. The identified change point from ES(t) is indicated by a black cross.
Following arrows: input training data representative of the ânormalâ dynamics is used to reconstruct the underlying phase space via a delay embedding (delayed components given by xi(t)). The constructed attractor network consists of a spatial and dynamics component, which discretises the dynamics of the system into a Markov chain. Hyperparameters ϵ, δ, Nmax and K determine the density of nodes and edges in the network (see âMethodsâ). Future incoming observations (test data) are compared against the expected transition probabilities in the attractor network and the level or surprise S(t) is calculated. This is subsequently used to calculate a normalised score ES(t), which determines the occurrence of change points.
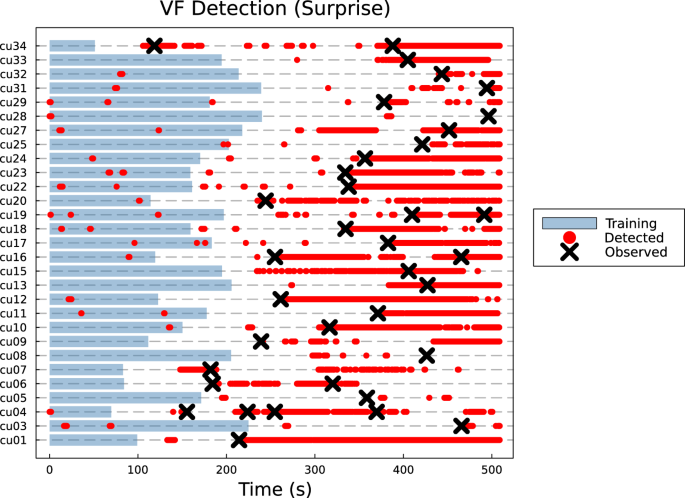
We demonstrate the effectiveness of attractor networks in a practical context by applying our method to the problem of automatic detection of ventricular fibrillation (VF) from electrocardiogram (ECG) time series. The single-channel ECG data is taken from the publicly available CU Ventricular Tachyarrhythmia data set on PhysioNet47,48. We find that the attractor networks approach is effective in detecting the onset of VF in 27 of 29 patients within 5âs of the annotated onset of VF. In 9 patients, the surprise measure S(t) was able to preempt the occurrence of VF (see Supplementary Note 1). We find that this approach outperforms competing statistical methods such as the moving average, standard deviation and permutation entropy16,49.
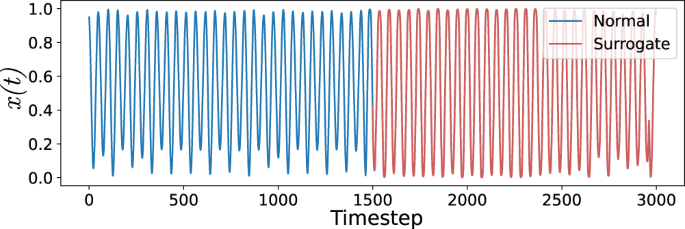
To further illustrate the flexibility of our approach, we test the effectiveness of attractor networks in detecting subtle dynamics changes in artificial time series. Specifically, we investigate if the surprise measures are able to discriminate between normal and surrogate data that possess the same statistical distributions. To do this, we artificially construct a time series consisting of alternating portions of normal and surrogate signals constructed from the Chua chaotic oscillator in the single scroll regime50. Surrogate data consists of artificially constructed, randomised data whose statistics and signal characteristics (e.g., power spectrum, mean, standard deviation) match those of the real reference data but are otherwise dynamically unrelated. These data are often used to perform tests for the existence of nonlinear and or deterministic dynamics in a given signal. In our analyses, we utilise surrogate data as a way to assess if our proposed attractor network is able to distinguish between time series with subtle dynamical differences but otherwise similar statistical structure. To do this, surrogate data are generated using the iterated amplitude-adjusted Fourier transform (AAFT) algorithm51. We find that our approach was able to reliably detect transitions between normal and surrogate time series and outperformed moving window approaches using statistical scores such as moving average, standard deviation and permutation entropy. We also provide results of additional change point detection tasks on other artificial time series in Supplementary Notes 2 and 3. Namely, (1) detecting subtle changes in phase space (phase coherence), and (2) detecting and quantifying gradual transitions and changes in system properties.
Results
Automated VF detection
ECG data are voltage and current measurements taken from the heartâs sinoatrial nodes, typically via externally placed electrodes. In a healthy regime, regular contraction of the heart to facilitate adequate blood circulation is driven by electrical impulses from the sinoatrial node. Each beat within a recorded ECG typically consists of three sections, P-QRS-T, corresponding to the movement of electrical wavefronts along the heart52.
ECG recordings contain complex information pertaining to various physiological dynamics. For example, the presence of cardiological irregularities and heart malfunction such as arrhythmia and tachycardia typically manifest as irregularities in an otherwise relatively regular pseudoperiodic signal (see Fig. 4). In this case, pseudoperiodicity can be attributed to minor fluctuations in the approximate frequency and amplitude of oscillations. More serious conditions such as atrial (AF) and ventricular fibrillation (VF) often result in the total degradation of the signal dynamics28. However, interpretation and analysis of ECG data are difficult and rely on trained medical professionals. This has led to a large body of methods that aim to automatically detect, interpret and diagnose physiological conditions from ECG data. These methods utilise analysis techniques from a wide range of disciplines such as statistics53,54, frequency analysis55,56, and machine learning57,58. For brevity, we refer interested readers to ref. 15 for a more comprehensive discussion of current ECG analysis methods.
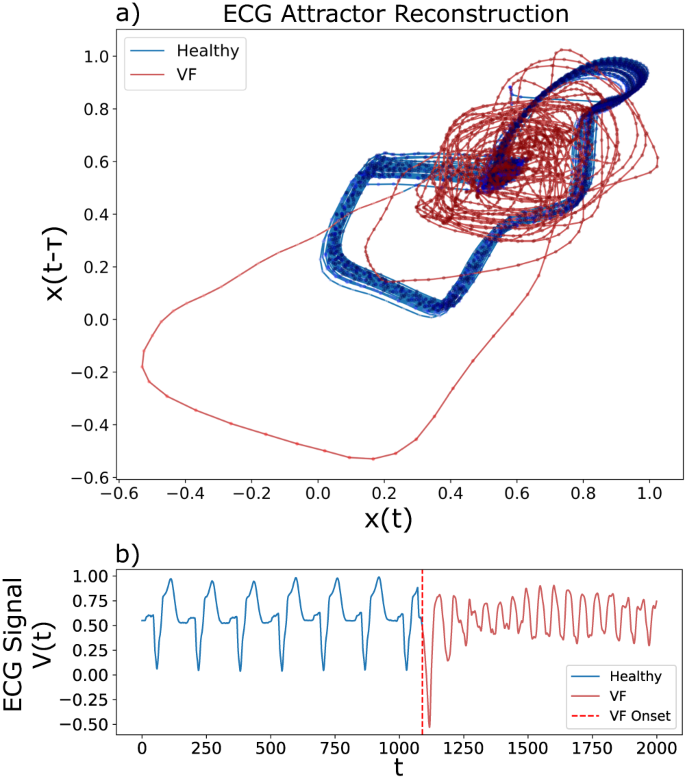
Healthy trajectories (blue) and ventricular fibrillation (VF) (red) shown. a Phase space reconstruction showing the trajectory of the healthy (blue) dynamics lying close to a stable orbit (the attractor). In contrast, the onset of VF (red) results in a structural collapse of the attractor. b An extract of the corresponding scalar ECG time series with the onset of VF labelled by a dashed vertical red line. The onset of VF is characterised by a change from regular oscillatory motions (blue) into erratic dynamics (red).
From the perspective of dynamical systems theory, healthy mode ECG can be equated to one that adheres well to some attractor (region in state space) that describes the healthy mode dynamics. The onset of VF causes stark changes in the characteristics of the ECG signal (see Fig. 4), which result in deviations from the original attractor. To demonstrate the utility of attractor networks, we apply the proposed method to the task of detecting ventricular fibrillation (VF) from an electrocardiogram (ECG) time series.
The CU Ventricular Tachyarrhythmia database47,48 consists of 8-min ECG recordings of 35 patients with eventual onset of VF. Each signal was recorded with a sampling rate of 250 Hz and filtered with a 70 Hz low-pass filter. The data set also contains annotations for each beat and the onset of VF. Of the 35 patients, 6 patients were excluded as there was either no clear annotation provided for the onset of VF, extended irregularities in the recorded ECG or the onset of VF was too close to the start of the recording. In most recordings, there were occasional occurrences of missing values most of which occur towards the end of the recording. These may be attributed to improper sensor placement or data logging. Generally, this does not heavily impact the performance of the detection algorithm as only one-step transitions are required to evaluate each surprise score. However, to prevent spurious false positives due to data dropouts, the surprise for transitions with missing values was set to 0.
A three-dimensional non-uniform delay embedding was used to reconstruct the phase space of the ECG dynamics. Delays were individually selected with the SToPS59 method based on the first 20,000 data points corresponding to the healthy dynamics prior to the onset of VF. The SToPS method works by scoring time scales based on how well their resulting 2D embeddings result in maximally circular holes in the resulting attractor. This is achieved by applying a delay embedding on randomly sampled short trajectory strands, computing its persistent homology and scoring each identified hole (1-dimensional homology) according to its circularity and its efficient use of points.
The attractor network was then constructed using the first half of the time prior to VF onset as training data. This training data was split equally between constructing the spatial and dynamics components of the network. The presence of both large and small-scale dynamics simultaneously within ECG results in a small, very high-density region when applying delay embedding, which can cause computational challenges. Therefore, a size parameter of ϵâ=â0.003 with a maximum of Nmaxâ=â6 nodes per observation was used to reduce computation time. The parameters ϵ and Nmax are used to control the density of the nodes and edges of the attractor network respectively (see âMethodsâ). For VF detection, S(t) and ES(t) of the remainder of each ECG recording including the VF onset are calculated. Change points are detected using thresholds S* and E* based on 95% confidence intervals calculated from S(t) and E(t) of the training data (see âMethodsâ). Exponential smoothing was applied with a characteristic time scale of 250 steps corresponding to 1âs of activity. Varying confidence levels effectively alter the level of specificity and sensitivity of the test. In our analyses, we select a confidence level of 95% due to its ubiquity, but this selection is arbitrary and may be adjusted depending on the needs of each application.
In order to quantify the methodâs VF detection performance, we define true positives as a detected change point that occurs within 5âs (1250 timesteps) before or after the first annotated onset of VF. Due to the nature of VF, the definition of false positives is not straightforward for subsequent VF episodes as the occurrence of VF can permanently alter the ECG dynamics even after recovery resulting in extended periods of detected change points well after the annotated VF onset. Any comparison against a threshold from previous âhealthyâ mode data is no longer relevant to the detection problem. We note that false positives can and do occur for times exceeding more than 5âs prior to the annotated onset of VF. However, from Fig. 5, these flags are relatively brief and are characteristically different to the persistent flags corresponding to a true VF episode.
For our analyses, we define two main metrics of performance for VF detection: (1) pâââ[0,â1] that measures the classification accuracy (healthy or VF) of the method near the onset of VF, and (2) pHâââ[0,â1] that is defined similarly to p but is weighted by the number of consecutive identical classifications (i.e., a string of classifications of VF will have a higher score than intermittent classifications) (see âMethodsâ for mathematical formulations). We also provide two additional performance metrics (see âMethodsâ section) that aim to quantify the performance of each method in preempting the onset of VF. This is accompanied by further discussion on the difficulties of quantifying VF detection performance in ECG data.
In both cases (p and pH), performance is measured across a time period 5âs before and after the first annotated onset of VF. Normalising over a time span of 10âs (5âs before and after VF), a score of pâ=â0.5 (or pH) generally corresponds to a perfect detection typically after the onset of VF. Scores larger than 1 generally correspond to detections occurring up to 5âs before the annotated onset of VF (i.e., preemptive detection).
A summary of detection performance for the four tested methods (moving average, moving standard deviation, moving permutation entropy and attractor network surprise) is given in Table 1. Moving statistics were calculated over sliding windows of 100 timesteps. The permutation entropy was calculated by converting the time series into a symbolic sequence with each sequence corresponding to an ordered sequence of observations ranked by magnitude16,49. This conversion utilised 7 uniform lags of size equal to the first lag calculated from SToPS59 in the delay embedding.
The final performance scores for each method are averaged over all 29 analysed patients. In our analyses, performance is assessed based on the successful detection of the first occurrence of VF. This is because following VF episodes, heart dynamics may persist for an extended period of time in a regime characterised by stress, which can alter the definition of the âhealthyâ mode. A more detailed breakdown of scores and comparison with other methods is given in Supplementary Note 4.
Overall, the attractor network surprise approach was able to more reliably detect and preempt the onset of VF compared to other methods based on moving statistics averaged across a sliding window. This is reflected in higher mean and median scores for p and pH as well. Focusing on individual cases, 25 out of the 29 patientsâ VF was detected within 5âs of the annotated onset (see Table 1, Fig. 5 and Supplementary Figs. S1 and S2 in Supplementary Note 1). Of the remaining four, three cases (cu05, cu08, cu28) of VF were not detected. The VF in the last case (cu25) was detected but was outside the 5-s window analysed. Surprisingly, 9 cases yielded detection scores greater than 1 and corresponded to the attractor network being able to preempt a plausible onset of VF. For 2 of these cases (cu13, cu15), the detections were made up to 3âmin before the annotated onset of VF (see Fig. 5). This may suggest that for some cases, subtle changes in ECG dynamics precede the onset of VF. For both cases where this was observed, these early detections corresponded to the increasing number and frequency of observed irregularities in the ECG relative to the healthy mode used for training. In almost all cases, false detections were found both within the training and testing portions of the ECG data. This is possibly due to dynamical irregularities in the time series such as cardiac arrhythmia or abnormal voltage amplitudes. However, most of these cases return a detection signal intermittently and are not persistent. This is in contrast to the onset of VF where the method persistently returns a positive result for detected change points.
Results are given for individual patients (IDs given by âcu..."). Annotated onsets for VF are given black crosses, and flagged detection of VF is given by a red dot. Blue regions represent the length of time series used for training and constructing the attractor network. The onset of VF is characterised by a persistent detection result (red).
Similar qualitative results were found for the other statistics-based detection methods. However, these methods slightly outperformed the attractor network approach in 9 of 29 patients. Of these, 3 patients (cu05, cu08, cu28) saw failures of almost all tested methods with the exception of the moving standard deviation and moving average performing slightly better for cu08 and cu28 respectively.
Amplitude-adjusted Fourier surrogates
The attractor network approach differs from those that track changes in moving statistics (e.g., CUSUM, Page-Hinkley Test, moving average test)25,60 in that detections are made based on observed differences in the dynamics of the system rather than the moving statistics of the observed time series. To investigate the effectiveness of the attractor network approach, we apply it to the task of distinguishing between changes in dynamical behaviour that partially preserve the stationary and non-stationary statistics of the data. This test can be formulated by artificially constructing a signal using conjoined portions of normal and amplitude-adjusted Fourier transform (AAFT) surrogate time series51. The result is an artificial data set that preserves the mean, standard deviation and power spectrum of the original data but lacks the original systemâs dynamical features. However, AAFT surrogates are unable to preserve the moving statistics of the signal. In our analyses, we select the Chua system operating in the single scroll regime that exhibits oscillatory dynamics that are relatively regular.
For change point detection, the attractor network was trained on 20,000 time steps in the regular single scroll Chua oscillator50, with an additional 20,000 time steps used to calculate threshold values for identifying change points. The test data consists of 7 concatenated portions of time series each of length 2000 steps alternating between the normal and surrogate data. Time series was generated by integrating the Chua equations with the RK4 algorithm and time step dtâ=â0.02. The equations are given by:
\(\\ f(y) =a{y}^{3}+by,\) where (β,âγ,âa,âb)â=â(53.612186, â0.75087096, 0.03755, â0.84154) and αâ=â17. Surrogates were generated using an iterated algorithm applied to a simulated Chua time series. The concatenations between normal and surrogate data ensured that endpoints were matched with no discontinuities. Direct observation of the scalar time series shows that differences between the surrogate and original data are not easily distinguishable (see Fig. 6). Permutation entropy was calculated with up to 7 lags, each a size equal to the first delay lag calculated using SToPS.
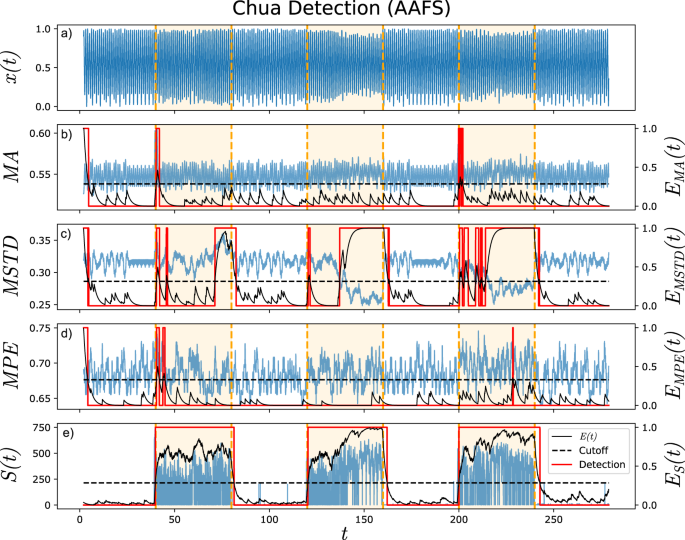
From Fig. 7, we find that the attractor network approach outperformed the moving statistics and permutation entropy. With the exception of the moving standard deviation, the remaining measures of moving average and permutation entropy fail (as expected) to detect changes in the time series.
From top to bottom: a Original time series, b moving average (MA), c moving standard deviation (MSTD), d moving permutation entropy (MPE), and e surprise scores S(t). Sliding window lengths of 100 steps were used for calculating moving averages. Detections (red) are made based on the exponential smoothed quantity E(t). Real change points are given by vertical orange lines. All comparison methods except MSTD struggle to reveal distinct changes in behaviour for the AAFT surrogates. In contrast, the attractor network approach is able to capture transitions well.
Sampling frequency effects
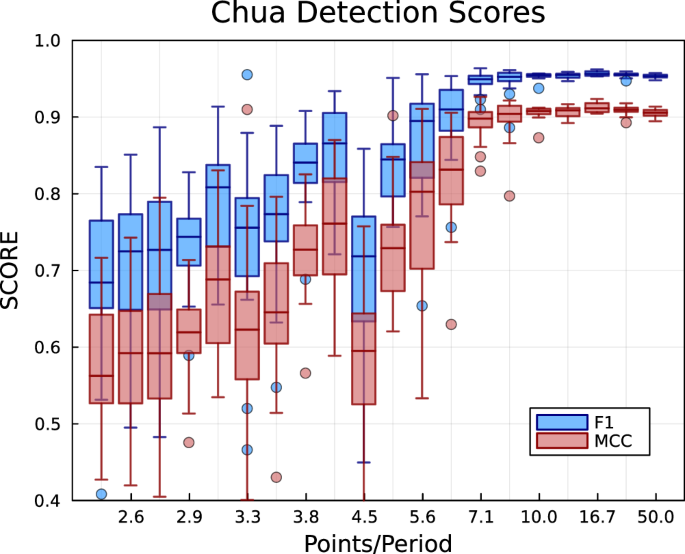
The attractor network effectively discretises the phase space dynamics into a Markov chain. Therefore, it is expected that the accuracy of the resulting attractor representation will be affected by the sampling frequency of the data. High sampling frequencies should not adversely affect the reconstruction as the attractor network construction algorithm iteratively simplifies dense cluster of points in phase space. The extent of this simplification is controlled by the size scale hyperparameter ϵ and δ (see âMethods: Spatial networkâ). However, input data points that are too sparse can limit the resolution of the attractor network and corresponding transitions. To test the effects of sampling, we construct attractor networks using input time series with varying sampling frequencies, which are then used to identify alternating transitions between the original and AAFT surrogate data. For this test, the analysed time series consisted of 20 alternations between normal and surrogate data with each segment lasting approximately 40 oscillations (2000 time steps). Performance is measured by the correct classification of each observed data point as either normal or abnormal. The F1 and Matthewâs correlation coefficient (MCC) were used to identify the classification accuracy (normal vs. surrogate) in each case.
The attractor network approach was found to perform well for sampling frequencies as low as 7 points per period (see Fig. 8). However, there is a decrease in performance for further decreases in sampling frequency. This is expected as time series with very low sampling frequencies effectively decrease the resolution of the phase space discretisation. Additionally, the observed transitions must be inferred across larger jumps in phase space, which can make the detection of abnormal dynamics difficult. This is particularly true for chaotic dynamics where the uncertainty of the future states exponentially increases in time with respect to small uncertainties in the initial conditions.
F1 recall score and Matthewâs correlation coefficient (MCC) are given for the attractor network approach with varying sampling frequencies (points per period). Scores show a large decrease in performance for sampling frequencies below 7 points per period. Outliers are given by 1.5 times the interquartile range.
Conclusion
We present a network-based change point detection method that aims to quantify deviations from the underlying attractor manifold of the target system. Significant deviations from the underlying attractor are used to infer the presence of a change point in the time series. To achieve this, input time series recorded from the healthy regime is delay embedded to represent the system dynamics in reconstructed phase space. These observed transitions are then used to construct an attractor network consisting of two components: the spatial network and the dynamics network. The resulting attractor network effectively acts as a Markov-chain representation of the dynamics along the systemâs attractor. Change point detection is achieved by calculating the measures of surprise S(t) and E(t) for each new observation respective to the learned attractor network. Unexpected observations caused by changes in the underlying systemâs dynamics result in large persistent values of surprise measures, which are used to flag change points.
The proposed approach was used to automatically detect the onset of ventricular fibrillation (VF) from recorded patient ECG data. Data was taken from the CU Ventricular Tachyarrhythmia data set that is publicly available on PhysioNet. The attractor network method was found to be sensitive in detecting the onset of VF for 25 out of 29 patients. In two cases, our approach was able to detect the occurrence of irregularities in the ECG well before the annotated onset VF provided in the data set.
To further illustrate the flexibility of our method, we apply the attractor network approach to the task of identifying subtle changes in the dynamics of the time series. This is done by constructing an artificial time series consisting of alternating portions of normal and abnormal behaviour. Specifically, we test the ability of the attractor network to detect transitions between normal and AAFT surrogate data. The attractor network and relevant metrics S(t) and E(t) were sensitive in distinguishing between normal and surrogate time series. We also find that this performance is maintained even for moderately low sampling frequencies. We provide in Supplementary Note 3 section results of additional change point detection tests and comparisons of the performance of the attractor network approach against moving windows approaches using statistical scores such as moving average, overall standard deviation and permutation entropy.
Methods
Time delay embedding
One of the most commonly employed methods of reconstructing the phase space of a system from observed scalar time series is via the method of time delay embedding. Consider a multivariate dynamical system \(\dot{\overrightarrow{x}}(t)=\overrightarrow{F}(\overrightarrow{x})\) in state space X where only the first component x1(t) is observed. Takensâ theorem guarantees generically that a delay embedding reconstruction \(\overrightarrow{y}(t)=({x}_{1}(t),{x}_{1}(t-{\tau }_{1}),\ldots ,{x}_{1}(t-{\tau }_{n}))\) defined in space Y with appropriately chosen delays (Ï1,ââ¦,âÏn) will have dynamics that are homeomorphic to the true system dynamics in X. That is, there exists a homeomorphism Φ such that,
Attractor networks are constructed with respect to some ambient phase space. Hence, the first step for analysing observed time series is constructing an embedding to augment the dimension of the input data. In our analyses, we use a non-uniform delay embedding,
where x(t), \(\overrightarrow{x}(t)\) are the original time series and reconstructed delay vector respectively and (Ï1,â.â.â.â,âÏn) are selected time lags.
The non-uniform delay embedding method is a generalisation of the uniform delay embedding often used for phase space reconstruction59. The latter method is simpler in its formulation and only requires the selection of two hyperparameters, the delay lag Ï and embedding dimension m. The resulting delay vector is then constructed as:
The lag Ï is typically selected using a mutual information criterion61, followed by the selection of m using a false nearest neighbour criterion62. However, reconstruction with a single lag is not adequate for systems with multiple disparate time and spatial scales63. This may be remedied by selecting a small Ï and large m at the expense of creating an overly high-dimensional phase space, which may computationally hinder later analyses. In contrast, non-uniform embedding allows the selection of multiple time scales without unnecessarily increasing the embedding dimension m. However, this requires solving the non-trivial problem of selecting relevant embedding delays Ï1,ââ¦,âÏm.
There are multiple proposed algorithms for the selection of non-uniform embedding delays such as MDOP64, PECUZAL65 and the method by Garcia and Almeida66. However, the resultant lags do not usually agree between algorithms due to their differing notions of optimal delays. Additionally, the calculated lags typically do not bear a clear explainable relationship to the time scales within the observed time series.
To address this, we use the SToPS algorithm, a persistent homology-based algorithm proposed by Tan et al.59, that identifies dynamically relevant time scales from the recurrence behaviour of chaotic and non-stationary time series. Instead of outputting a collection of time lags, SToPS calculates the significance of each potential time lag in constructing a spectrum of time scales from which individual lag values may be selected to construct a delay vector. This provides a more accurate, explainable and robust method for analysing time series that may contain multiple disparate time scales and magnitudes, such as ECG, by allowing for the construction of well-unfolded attractors that more accurately capture the underlying dynamics in phase space.
Such an approach is reminiscent of a Fourier power spectrum, with the added benefit of being applicable to data with chaotic behaviour or non-stationary statistics.
Phase space reconstruction to attractor networks
Following the time delay embedding of time series to reconstruct the systemâs attractor in phase space, the next step requires the construction of attractor networks by discretising phase space. This step involves the construction of two components: the spatial and dynamics network.
Spatial network
One of the drawbacks of the network-based method for identifying attractors presented by St Luce et al.33 is the potentially poor computational scaling for increasing phase space dimension. This is due to the grid-based voxel scheme used to discretise the entire phase space, where each voxel is represented by a single node33. This results in a prohibitively large network for even modest phase space dimensions that require extensive pruning before any further computation can be done.
To address this, we propose instead to use the original positions of the input data to guide the discretisation of phase space. Randomly sampled points from the embedded training data are selected as kernels in phase space and used to construct a sparse point cloud representation of the attractor. Additional sampled points are then progressively added in cycles until the desired density or number of points in the attractor is reached. However, depending on the density distribution of the attractor and sampling of points, the simple addition of more points may not always increase the detail of the attractor structure captured by the point cloud. This results from the presence of redundant points that are near neighbours in high-density regions of the attractor. To address this, the point cloud is trimmed by replacing a high-density cluster of points in the attractor with a single âcentre-of-massâ point. The algorithm for iteratively constructing the spatial network is given as follows:
-
Let A be the collection of points in the spatial network and ϵ be a predefined size scale.
-
Sample a set of points S from the training input data set and add it to A: \((S\cup A)\to A\)
-
Calculate the pairwise distance δij for all point pairs \(({\overrightarrow{x}}_{i},{\overrightarrow{x}}_{j})\in A\). Define a neighbour adjacency matrix D with entries:
$${D}_{ij}=\left\{\begin{array}{ll}1,\quad &0\, < \,{\delta }_{ij}\, < \,\epsilon \\ 0,\quad &{\delta }_{ij}\, > \,\epsilon \hfill \end{array}\right.$$(4) -
For every node i in the network represented by D, calculate the local clustering coefficient ci,
$${c}_{i}=\frac{2\,{n}_{\Delta i}}{{k}_{i}({k}_{i}-1)},$$(5)where ki and nÎi are the degree and number of triangles passing through node i respectively.
-
Let C be the collection of nodes whose clustering coefficient ciâ>â0.5, and kmax be the highest degree amongst all the nodes in C. Identify \({C}_{{k}_{max}}\) corresponding to the set of all nodes with degree kmax.
-
For each \({\overrightarrow{x}}_{i}\in {C}_{{k}_{max}}\) with kmax neighbours, calculate the average location across its kmax neighbours \({\overrightarrow{x}}_{j}\) given by,
$${\overrightarrow{x}}_{i}^{* }=\frac{1}{{k}_{max}}\mathop{\sum }\limits_{j=1}^{{k}_{max}}{\overrightarrow{x}}_{j},$$(6) -
Replace the cluster of points \({C}^{{\prime} }\) in A with the average point, \((A\setminus {C}_{{k}_{max}})\cup \{{\overrightarrow{x}}_{i}^{* }\}\to A\), where ⧹ corresponds to the set difference operator.
-
Repeat steps 3 and 7 until kmaxâ<â3 (i.e., no triangle clusters exist).
-
Repeat steps 2 to 6 until all the input training data has been processed.
In the above formulation, the density parameter ϵ prescribes a minimum allowable distance between point pairs in the attractor. This limits the amount of redundant points in high-density regions, which alleviates the computational burden in later steps. The cutoff threshold for the maximum clustering may also be adjusted to control the aggressiveness of the trimming procedure. The algorithm is also self-terminating as the number of nodes added in each iteration will always be less than or equal to the number of nodes removed in Step 7.
Dynamics network
The dynamics network aims to describe the vector field dynamics of the system within phase space. However, this first requires a discretisation of continuous phase space. To do this, the points in A are used as kernels to discretise the continuous phase space into a finite collection of bounded cells with characteristic spatial scale δ. Because A only contains points that are relevant to the attractor of the system, the resulting discretisation automatically excludes regions which are greater than a notional distance of δ from the attractor. This removes the need to trim off irrelevant regions of the discretised phase space, previously required in the grid-based voxel approach by St Luce et al.33.
In order to encode vector fields and dynamics of trajectories into the attractor network representation, pairs of observed transitions between successive points in phase space are used to create a transition matrix.
-
Let M be an â£Aâ£âÃââ£A⣠matrix, B is the set of input training points for learning the dynamics network and A is the collection of points in the spatial network.
-
Consider a point \({\overrightarrow{x}}_{i}(t)\in B\) and its observed next position \({\overrightarrow{x}}_{i}(t+dt)\). The pair of points \(({\overrightarrow{x}}_{i}(t),{\overrightarrow{x}}_{i}(t+dt))\) correspond to a transition between two regions of the discretised state space.
-
Identify up to Nmax each of attractor points \(\overrightarrow{a},\overrightarrow{b}\in A\) that are within a distance δ from \(({\overrightarrow{x}}_{i}(t),{\overrightarrow{x}}_{i}(t+dt))\) respectively. The hyperparameter Nmax controls the edge density of the resultant attractor network. Calculate the corresponding distances δa,âδb where,\({\delta }_{a}=\parallel {\overrightarrow{x}}_{i}(t)-\overrightarrow{a}\parallel ,\\ {\delta }_{b}=\parallel {\overrightarrow{x}}_{i}(t+dt)-\overrightarrow{b}\parallel.\)
-
Calculate a scaling value α given by the following expression,
$$\alpha =f(\delta )=\left\{\begin{array}{ll}\sqrt{{\left(\frac{{\delta }_{a}}{\delta }\right)}^{2}+{\left(\frac{{\delta }_{b}}{\delta }\right)}^{2}}, &{\delta }_{a},{\delta }_{b}\, < \,\delta \\ 0,\hfill &\,{{\mbox{otherwise}}}\,\end{array}\right.$$(7) -
Add a corresponding weight to the element Mab corresponding the transition between attractor points \(\overrightarrow{a},\overrightarrow{b}\in A\),\({M}_{ab}+{e}^{-\alpha K}\to {M}_{ab},\) where K is a shape parameter.
-
Repeat steps 2â5 for all points \({\overrightarrow{x}}_{i}(t)\in B\).
Observed transitions in the training data are used to record the frequency of transitions between any two points in discretised phase space. The characteristic spatial scale δ is taken to be the 99% quantile of all closest neighbour distances from points included in the spatial network. The value α captures the goodness of fit between the endpoints of an observed transition and the available possible attractor points in the discretised space. Lower values of α result in a contribution closer to 1 to the tally of transitions. For αâ>â0, inaccuracies in the fit are scaled by K, which governs the penalty applied when calculating the contribution of the observed transition.
Finally, the matrix M may be converted into a stochastic transition matrix Mf, which we label the flow matrix. Combined, flow matrix Mf and attractor points A work together to form a discretised network representation of system dynamics in phase space where Mf is likened to the Perron-Frobenius operator acting on a domain A.
One additional consideration is that the directed matrix M will likely have nodes with no outdegree. These correspond to points in A that are infrequently visited or are strong sinks or stable nodes. Therefore, calculating Mf requires the iterative removal of all nodes with 0 outdegree until none remain.
Surprise!
The phase space approach to change point detection relies on the argument that changes in the underlying dynamics of the data-generating process correspond to trajectories in phase space that deviate from the original attractor of the system when operating in its normal state. In terms of the discretised phase space, this would correspond to the observation of transitions that are either unlikely or not previously encountered within the training set. The attractor network approach to change point detection uses the calculation of a metric S(t), named the âsurpriseâ, which aims to quantify the level of surprise observed in a given transition.
Consider an observed transition at time t between two nodes iâââj in the constructed attractor network. Let ki be the outdegree of node i, and pij be the calculated probability of transition from the stochastic flow matrix Mf. Let N be the number of data points used to train the attractor network. If a transition between nodes iâââj was not observed within N observations, pij is assigned a value of 1/2N to avoid singularities,
Let Hmax(i) be the maximum possible entropy for a given node i in the attractor network based on its outdegree kmax. This corresponds to the case where all outdegree transitions are equally probable,
Similarly, H(i) is defined as the actual entropy calculated based on the probability distribution of all ki outdegrees for node i,
The quantification of surprise requires a normalisation with respect to the maximum possible entropy for each source node i to account for variations in the outdegree across different nodes in the attractor network. Observed uncommon transitions in uniformly distributed, high outdegree nodes are less informative as each outcome is equally unlikely. Therefore, we define a weighting value, η, to quantify the meaningfulness of an observation with respect to the expected potential transitions in the attractor network,
The value of surprise S(t)â=âS(i,âj) corresponding to an observed transition between two nodes i,âj at time t in the constructed attractor network is then given by the normalised expression,
Change point detection
One common approach to change point detection is to identify deviation in a statistical measure between a known healthy mode and incoming observations. Significant deviations in this measure can be used to infer the presence of a change point. For example, the attractor network approach for change point detection corresponds to identifying consecutive transitions with persistently high values of surprise (i.e., trajectories are more frequently deviating away from the attractor of the normal state).
More generally, given some measured statistic S(t) (such as the attractor network surprise), it is possible to convert S(t) into a binary measure of normal vs. abnormal by filtering according to some interval [Smin,âSmax]. Therefore, we can define a new binary measure \(\hat{S}(t)\),
where \(\hat{S}(t)=1\) corresponds to an observation that is abnormal. The values of Smin and Smax can be selected as quantiles of the distribution of observed S(t) from data that is known to be in the healthy regime (i.e., from the training data set). In the case of attractor network surprise, (Smin,âSmax)â=â(0,âS*) where S* is selected to be the 95% upper quantile of the data because surprise is always positive.
Depending on the statistic, temporary and benign fluctuations in the tracked measure S(t) may result in brief, intermittent classifications of observations as abnormal (i.e., \(\hat{S}(t)=1\)). To ensure that an observed change is indeed a genuine change point, incoming observations must be consistently classified as abnormal for an extended period of time ÏE to be flagged as potential change points. This can be done by applying exponential smoothing to the binary series \(\hat{S}(t)\),
where βâ=â1/ÏE is a smoothing parameter. For automatic change point detection, change points are defined when E(t) exceeds some threshold kââ âE*, where E* is the 95-percentile of observed E(t) in the input training data set and k is a multiplier term that determines the strength of the threshold. Because E(t) can be calculated with respect to an arbitrary measure S(t), we use the following notation EMA(t), EMSTD(t), EMPE(t) and ES(t) for the exponential smoothed series resulting from the moving average, moving standard deviation, moving permutation entropy and attractor network surprise scores respectively.
Performance quantification
Detection scores p and p H
To quantify the accuracy of detecting VF onset, we propose two measures of the true positive rate given p and pH. A true positive detection is defined as the classification of an observation as abnormal for a time period [ts,âte] before or after the annotated onset of VF with a total length T timesteps.
The first measure of performance p is calculated as the proportion of timesteps within the abovementioned time period that are correctly classified as abnormal.
The second measure of performance pH is a modified form of p that accounts for both the proportion of true positive classifications and the persistence of the classification. In this measure, a string of identical classifications would be weighted higher than those with intermittent true positives. The score for pH is calculated as,
where H is the normalised entropy of the distribution of the ordered observations given by,
The probability pn is calculated by partitioning the observed string of classifications into groups of successively identical classifications. For example, consider an observed time period with 10 timesteps with a string of classifications given by 0001101111 where a value of 1 corresponds to an observation that is classified as abnormal. This set of observations may be partitioned into 4 groups resulting in the sets of probabilities (p1,âp2,âp3,âp4)â=â(0.3,â0.2,â0.1,â0.4). Therefore, the quantity H is a measure of the fragmentation of an observed string of classifications where larger values of H indicate more fragmented and intermittent classification of observations of either normal or abnormal.
Quantifying preemptive detection performance
One of the desirable qualities of online change point detection is to predict the occurrence of a change point for some period of time before an event. This can be particularly useful in mission critical systems, such as VF detection, where early or preemptive detection can inform appropriate intervention measures. In the context of VF detection, it is common for arrhythmias to occur prior to the onset of VF. This occurrence potentially suggests gradual changes in the heart dynamics prior to VF, which may otherwise be difficult to detect by visual inspection of the ECG. The detection of this dynamical change may be used to inform the early warning detection of VF.
For our VF detection analyses, we propose two different measures to quantify and compare the ability of each change point detection method in preempting the onset of VF. For both measures, we consider a time interval [tVFâââÏp,âtVF] of increasing length Ïp prior to the annotated onset of VF. For every given Ïp, we calculate two measures across the time interval: (1) the length of the longest uninterrupted streak of positive detections leading up to the onset of VF, and (2) the proportion of positive detections across the entire time interval. The first measure aims to measure the degree of persistence in the positive detection rate where longer positive detection streaks suggest a more confident preemptive detection of an incoming VF episode, whereas the second measure is a naïve measure of the positive detection rate.
Due to the nature of the data and lack of information regarding the real physiological condition of each patient, it is not possible to determine if early detections from each method are true or false positives. Therefore in both cases, we assume that all positives are true positive and limit our analyses to only a relatively short maximum window length Ïpâ=â1250 prior to the annotated onset of VF. Only detection relating to the first annotated VF onset in each patient is used to calculate the results as physiological changes to the heart following VF may result in permanent changes to ECG dynamics, which may yield an artificially increased number of positive detections.
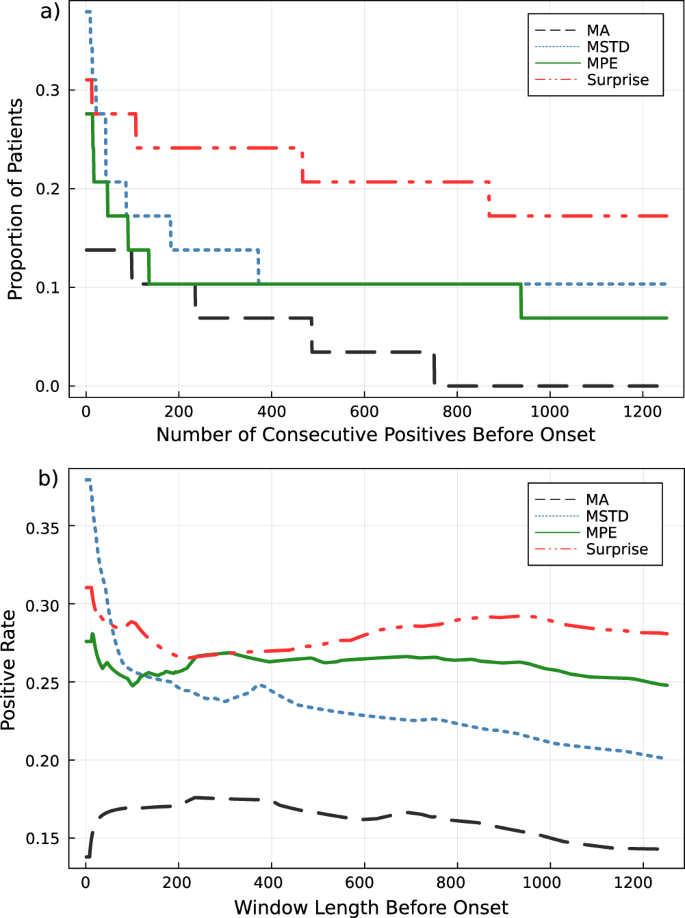
Scores are calculated for each individual patient. A final performance score for each detection method is then taken as the average scores across all 29 analysed patient ECGs (see Fig. 9a, b). The attractor network surprise approach was found to provide a longer and more persistent streak of positive detection leading up to the onset of VF compared to the moving average, standard deviation and permutation entropy methods. Positive detection rates were also slightly higher for the attractor network surprise method.
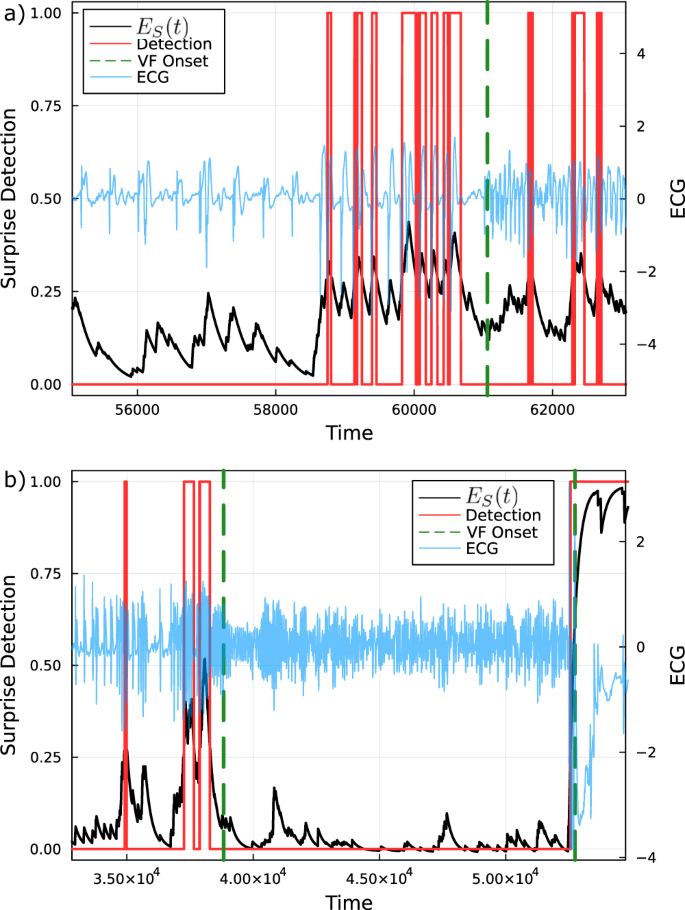
In Fig. 9b, it is expected that increasing window lengths will cause gradual decreases in the positive rate as the window length begins to include periods of normal healthy behaviour. However, we find that for the attractor network surprise and moving standard deviation methods, there is a peak in the performance rate partway through the profile. Further inspection into the failure modes of each patient reveals that this is the result of the method detecting the occurrence of irregular ECG behaviour for some period before the annotated onset of VF (see Fig. 10). At the onset, some of these signals collapse into high-frequency oscillatory dynamics. This can correspond to a trajectory that lingers in a high-density region of the attractor network resulting in artificially low suprise values.
a Patient cu20 exhibits irregular oscillatory dynamics for an extended period prior to the annotated onset that is detected by the attractor network. However, the signal collapses into a high-frequency oscillation following onset. b Patient cu04 where signal irregularities are detected before the onset of VF (shown by the two peaks in the surprise) followed by a similar collapse into high-frequency oscillatory dynamics. The second VF in the ECG recording is subsequently detected later.
Code availability
All code pertaining to the above analyses are publicly available on the online GitHub repository at https://github.com/eugenetkj98/AttractorNetworksPublic.
References
Goswami, B. et al. Abrupt transitions in time series with uncertainties. Nat. Commun. 9, 48 (2018).
Majdandzic, A. et al. Multiple tipping points and optimal repairing in interacting networks. Nat. Commun. 7, 10850 (2016).
Boers, N., Marwan, N., Barbosa, H. M. & Kurths, J. A deforestation-induced tipping point for the South American monsoon system. Sci. Rep. 7, 41489 (2017).
Boers, N. & Rypdal, M. Critical slowing down suggests that the western Greenland Ice Sheet is close to a tipping point. Proc. Natl Acad. Sci. USA 118, e2024192118 (2021).
Jiang, J. et al. Predicting tipping points in mutualistic networks through dimension reduction. Proc. Natl Acad. Sci. USA 115, E639âE647 (2018).
Nelson, B., McGorry, P. D., Wichers, M., Wigman, J. T. & Hartmann, J. A. Moving from static to dynamic models of the onset of mental disorder: a review. JAMA Psychiatry 74, 528â534 (2017).
Trefois, C., Antony, P. M., Goncalves, J., Skupin, A. & Balling, R. Critical transitions in chronic disease: transferring concepts from ecology to systems medicine. Curr. Opin. Biotechnol. 34, 48â55 (2015).
Staudacher, M., Telser, S., Amann, A., Hinterhuber, H. & Ritsch-Marte, M. A new method for change-point detection developed for on-line analysis of the heart beat variability during sleep. Phys. A 349, 582â596 (2005).
Chen, G., Lu, G., Shang, W. & Xie, Z. Automated change-point detection of EEG signals based on structural time-series analysis. IEEE Access 7, 180168â180180 (2019).
Schröder, A. L. & Ombao, H. Fresped: frequency-specific change-point detection in epileptic seizure multi-channel EEG data. J. Am. Stat. Assoc. 114, 115â128 (2019).
Donges, J. F. et al. Nonlinear detection of paleoclimate-variability transitions possibly related to human evolution. Proc. Natl Acad. Sci. USA 108, 20422â20427 (2011).
Ducré-Robitaille, J.-F., Vincent, L. A. & Boulet, G. Comparison of techniques for detection of discontinuities in temperature series. Int. J. Climatol. 23, 1087â1101 (2003).
Gallagher, C., Lund, R. & Robbins, M. Changepoint detection in climate time series with long-term trends. J. Clim. 26, 4994â5006 (2013).
Thorne, B., Jüngling, T., Small, M., Corrêa, D. & Zaitouny, A. Reservoir time series analysis: using the response of complex dynamical systems as a universal indicator of change. Chaos 32, 033109 (2022).
Aminikhanghahi, S. & Cook, D. J. A survey of methods for time series change point detection. Knowl. Inf. Syst. 51, 339â367 (2017).
Unakafov, A. M. & Keller, K. Change-point detection using the conditional entropy of ordinal patterns. Entropy 20, 709 (2018).
Zameni, M. et al. Unsupervised online change point detection in high-dimensional time series. Knowl. Inf. Syst. 62, 719â750 (2020).
Reddy, S. et al. Using mobile phones to determine transportation modes. ACM Trans. Sens. Netw. 6, 1â27 (2010).
Desobry, F., Davy, M. & Doncarli, C. An online kernel change detection algorithm. IEEE Trans. Signal Process. 53, 2961â2974 (2005).
Jin, B., Chen, Y., Li, D., Poolla, K. & Sangiovanni-Vincentelli, A. A one-class support vector machine calibration method for time series change point detection. In Proc. 2019 IEEE International Conference on Prognostics and Health Management (ICPHM), 1â5 (IEEE, 2019).
Klinkenberg, R. & Joachims, T. Detecting concept drift with support vector machines. In Proc. 17th International Conference on Machine Learning, 487â494 (2000).
Cleland, I. et al. Evaluation of prompted annotation of activity data recorded from a smart phone. Sensors 14, 15861â15879 (2014).
Saatçi, Y., Turner, R. D. & Rasmussen, C. E. Gaussian process change point models. In Proc. 27th International Conference on Machine Learning (ICML-10), 927â934 (2010).
Adams, R. P. & MacKay, D. J. Bayesian online changepoint detection. Preprint at arXiv https://doi.org/10.48550/arXiv.0710.3742 (2007).
da Costa, F. G., Rios, R. A. & de Mello, R. F. Using dynamical systems tools to detect concept drift in data streams. Expert Syst. Appl. 60, 39â50 (2016).
Abarbanel, H. Analysis of Observed Chaotic Data (Springer, 2012).
Kantz, H. & Schreiber, T. Nonlinear Time Series Analysis, Vol. 7 (Cambridge University Press, 2004).
Roopaei, M., Boostani, R., Sarvestani, R. R., Taghavi, M. A. & Azimifar, Z. Chaotic based reconstructed phase space features for detecting ventricular fibrillation. Biomed. Signal Process. Control 5, 318â327 (2010).
Vemishetty, N. et al. Phase space reconstruction based CVD classifier using localized features. Sci. Rep. 9, 14593 (2019).
Marwan, N., Romano, M. C., Thiel, M. & Kurths, J. Recurrence plots for the analysis of complex systems. Phys. Rep. 438, 237â329 (2007).
Huang, N. E. et al. The empirical mode decomposition and the Hilbert spectrum for nonlinear and non-stationary time series analysis. Proc. R. Soc. Lond. A Math. Phys. Eng. Sci. 454, 903â995 (1998).
Takens, F. Detecting strange attractors in turbulence. Dynamical Systems and Turbulence, Warwick 1980. In Proc. Symposium Held at the University of Warwick 1979/80, 366â381 (Springer, 2006).
St Luce, S. & Sayama, H. Analysis and visualization of high-dimensional dynamical systemsâ phase space using a network-based approach. Complexity 2022, 3937475 (2022).
Zhang, J. & Small, M. Complex network from pseudoperiodic time series: topology versus dynamics. Phys. Rev. Lett. 96, 238701 (2006).
Donner, R. V., Zou, Y., Donges, J. F., Marwan, N. & Kurths, J. Recurrence networks-a novel paradigm for nonlinear time series analysis. N. J. Phys. 12, 033025 (2010).
McCullough, M., Small, M., Stemler, T. & Iu, H. H.-C. Time lagged ordinal partition networks for capturing dynamics of continuous dynamical systems. Chaos 25, 053101 (2015).
Zhang, J. et al. Constructing ordinal partition transition networks from multivariate time series. Sci. Rep. 7, 7795 (2017).
Xu, X., Zhang, J. & Small, M. Superfamily phenomena and motifs of networks induced from time series. Proc. Natl Acad. Sci. USA 105, 19601â19605 (2008).
Lacasa, L., Luque, B., Ballesteros, F., Luque, J. & Nuno, J. C. From time series to complex networks: the visibility graph. Proc. Natl Acad. Sci. USA 105, 4972â4975 (2008).
Myers, A., Munch, E. & Khasawneh, F. A. Persistent homology of complex networks for dynamic state detection. Phys. Rev. E 100, 022314 (2019).
Emrani, S., Gentimis, T. & Krim, H. Persistent homology of delay embeddings and its application to wheeze detection. IEEE Signal Process. Lett. 21, 459â463 (2014).
Khasawneh, F. A. & Munch, E. Chatter detection in turning using persistent homology. Mech. Syst. Signal Process. 70, 527â541 (2016).
Islambekov, U., Yuvaraj, M. & Gel, Y. R. Harnessing the power of topological data analysis to detect change points. Environmetrics 31, e2612 (2020).
Ofori-Boateng, D., Dominguez, I. S., Akcora, C., Kantarcioglu, M. & Gel, Y. R. Topological anomaly detection in dynamic multilayer blockchain networks. Machine Learning and Knowledge Discovery in Databases. Research Track: European Conference, ECML PKDD 2021, 788â804 (Springer, 2021).
Xie, L. & Xie, Y. Sequential change detection by optimal weighted l2 divergence. IEEE J. Sel. Areas Inf. Theory 2, 747â761 (2021).
Zheng, X., Mak, S., Xie, L. & Xie, Y. Percept: a new online change-point detection method using topological data analysis. Technometrics 65, 162â178 (2023).
Goldberger, A. L. et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101, e215âe220 (2000).
Nolle, F., Badura, F., Catlett, J., Bowser, R. & Sketch, M. CREI-GARD, a new concept in computerized arrhythmia monitoring systems. Comput. Cardiol. 13, 515â518 (1986).
Cao, Y., Tung, W. W., Gao, J., Protopopescu, V. A. & Hively, L. M. Detecting dynamical changes in time series using the permutation entropy. Phys. Rev. E 70, 046217 (2004).
Kengne, J. On the dynamics of Chuaâs oscillator with a smooth cubic nonlinearity: occurrence of multiple attractors. Nonlinear Dyn. 87, 363â375 (2017).
Schreiber, T. & Schmitz, A. Surrogate time series. Phys. D 142, 346â382 (2000).
Berkaya, S. K. et al. A survey on ECG analysis. Biomed. Signal Process. Control 43, 216â235 (2018).
Song, C., Liu, K., Zhang, X., Chen, L. & Xian, X. An obstructive sleep apnea detection approach using a discriminative hidden Markov model from ECG signals. IEEE Trans. Biomed. Eng. 63, 1532â1542 (2015).
Dilmac, S. & Korurek, M. ECG heart beat classification method based on modified ABC algorithm. Appl. Soft Comput. 36, 641â655 (2015).
Castillo, E. et al. Efficient wavelet-based ECG processing for single-lead FHR extraction. Digit. Signal Process. 23, 1897â1909 (2013).
Pinto, J. R., Cardoso, J. S. & Lourenço, A. Evolution, current challenges, and future possibilities in ECG biometrics. IEEE Access 6, 34746â34776 (2018).
Alonso-Atienza, F., Morgado, E., Fernandez-Martinez, L., Garcia-Alberola, A. & Rojo-Alvarez, J. L. Detection of life-threatening arrhythmias using feature selection and support vector machines. IEEE Trans. Biomed. Eng. 61, 832â840 (2013).
Li, Q., Rajagopalan, C. & Clifford, G. D. Ventricular fibrillation and tachycardia classification using a machine learning approach. IEEE Trans. Biomed. Eng. 61, 1607â1613 (2013).
Tan, E. et al. Selecting embedding delays: an overview of embedding techniques and a new method using persistent homology. Chaos 33, 032101 (2023).
Page, E. S. Continuous inspection schemes. Biometrika 41, 100â115 (1954).
Fraser, A. M. & Swinney, H. L. Independent coordinates for strange attractors from mutual information. Phys. Rev. A 33, 1134 (1986).
Kennel, M. B. & Abarbanel, H. D. False neighbors and false strands: a reliable minimum embedding dimension algorithm. Phys. Rev. E 66, 026209 (2002).
Judd, K. & Mees, A. Embedding as a modeling problem. Phys. D 120, 273â286 (1998).
Nichkawde, C. Optimal state-space reconstruction using derivatives on projected manifold. Phys. Rev. E 87, 022905 (2013).
Krämer, K. H. et al. A unified and automated approach to attractor reconstruction. N. J. Phys. 23, 033017 (2021).
Garcia, S. P. & Almeida, J. S. Nearest neighbor embedding with different time delays. Phys. Rev. E 71, 037204 (2005).
Acknowledgements
E.T. is supported by a Robert and Maude Gledden Postgraduate Research Scholarship and the Australian Government RTP Scholarship at The University of Western Australia. M.S. and D.C. acknowledge the support of the Australian Research Council through the Centre for Transforming Maintenance through Data Science (grant number IC180100030), funded by the Australian Government. M.S. is also supported by the ARC Discovery Grant DP200102961, funded by the Australian Government.
Author information
Authors and Affiliations
Contributions
E.T. conceived the ideas for this project and provided implementations for the algorithms. S.D.A., D.C., M.S., and T.S. provided supervision and concept refinement. The manuscript was prepared by E.T. with reviews and revisions provided by S.D.A., D.C., M.S., and T.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Physics thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisherâs note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the articleâs Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the articleâs Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tan, E., Algar, S.D., Corrêa, D. et al. Network representations of attractors for change point detection. Commun Phys 6, 340 (2023). https://doi.org/10.1038/s42005-023-01463-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42005-023-01463-y
This article is cited by
-
Symmetrized Dot Pattern as an Alternative Method to Visualize the Dynamics of Tokamak Plasma Radiation
Journal of Fusion Energy (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.