Abstract
Next-generation sequencing (NGS) is currently the method of choice for analyzing gut microbiota composition. As gut microbiota composition is a potential future target for clinical diagnostics, it is of utmost importance to enhance and optimize the NGS analysis procedures. Here, we have analyzed the impact of DNA extraction and selected 16S rDNA primers on the gut microbiota NGS results. Bacterial DNA from frozen stool specimens was extracted with 5 commercially available DNA extraction kits. Special attention was paid to the semiautomated DNA extraction methods that could expedite the analysis procedure, thus being especially suitable for clinical settings. The microbial composition was analyzed with 2 distinct protocols: 1 targeting the V3–V4 and the other targeting the V4–V5 area of the bacterial 16S rRNA gene. The overall effect of DNA extraction on the gut microbiota 16S rDNA profile was relatively small, whereas the 16S rRNA gene target region had an immense impact on the results. Furthermore, semiautomated DNA extraction methods clearly appeared suitable for NGS procedures, proposing that application of these methods could importantly reduce hands-on time and human errors without compromising the validity of results.
Keywords: next-generation sequencing, fecal samples, sample preprocessing, microbial diversity
INTRODUCTION
The human gastrointestinal tract harbors an extensively diverse microbial ecosystem that has an important role in the health and physiologic functions of the host.1, 2 For instance, gut microbes maintain gut barrier function, take part in food digestion, and regulate immune functions.2, 3 Gut microbiota dysbiosis, referring to an aberrant gut microbiota composition, has been linked to several diseases and disorders, such as obesity, diabetes, and inflammatory bowel disease.4–6 Therefore, the studying of taxonomic-level associations of gut microbiota with different diseases is currently of great interest, and gut microbiota composition is an interesting potential future target for clinical diagnostics.7
In the past decades, molecular techniques targeting the bacterial 16S rRNA gene or other genetic markers have remarkably advanced the study of microbial communities.8, 9 These methods have, in a significant degree, replaced the traditional culture-based techniques that are eminently inadequate for comprehensive gut microbiota studies; it has been estimated that still today, less than one-half of the gut microbes is cultivable with the standard laboratory protocols.1, 10 Molecular methods do not require viable bacteria and are thus capable of providing a more comprehensive view of the microbial community structure of the gut.11, 12 The selection of available molecular methods is relatively broad, but in recent years, NGS has revolutionized the gut microbiota research.11–13 NGS technologies enable fast and high-throughput analyses, and as the methodological advancements have led to a significant decrease in the analysis costs, NGS has become a feasible and compelling method for studying intestinal microbiota.13–15 For example, in bacterial 16S rRNA gene-targeting studies, NGS enables a high-throughput analysis of the gut microbiota at a very reasonable cost, and it has recently greatly expanded the knowledge of this complex ecosystem.8, 13, 16 However, since the number of available NGS methods is extensive, and the optimization of the methods is demanding, an unbiased comparison of different studies is still a challenge.9 For example, DNA extraction has been proposed to influence the outcome of microbiota studies,17–19 and furthermore, targeted 16S rRNA gene variable regions have been shown to affect the results significantly.20–25 In addition, interpretation of the sequencing data is challenging and demands bioinformatic specialists.26
As high-throughput capacity is one of the major advantages of NGS techniques, it is surprising that the DNA extraction methods applied are often manual and time-consuming. In fact, DNA extraction can be considered as a bottleneck of otherwise high-throughput microbiota NGS studies. Even the DNA extraction methods recommended by the International Human Microbiome Standards (IHMS) project (2011–2015)27 are completely manual, i.e., require a substantial amount of hands-on time and are thus not suitable for large-scale studies or routine diagnostics in clinical microbiology laboratories. Furthermore, manual protocols are always prone to human errors that may lead to significant variation among experiments. Therefore, evaluation of the suitability of less time-demanding procedures for DNA extraction for microbiota studies is necessary, as these methods would be more suitable to be incorporated in routine diagnostic settings. In a study by Claassen et al.,28 an automatic DNA extraction method has been found more effective than manual methods for quantitative PCR methods. However, automatic DNA extraction methods have not, to our knowledge, been previously evaluated for high-throughput 16S rRNA gene sequencing. Here, we have thus evaluated the suitability of 2 semiautomated DNA extraction procedures that could be easily assimilated in large-scale studies or diagnostic laboratory routine. In summary, to improve the quality and accuracy of the NGS-based microbiota analyses, we have studied the impact of DNA extraction methods and selected 16S rDNA primers on the results of the gut microbiota composition analysis, paying special attention to the semiautomated DNA extraction protocols.
MATERIALS AND METHODS
Fresh stool specimens from 4 adult donors were derived from a human study that was approved by the Ethics Committee of the Central Finland Health Care District. Written, informed consent was obtained from each of the 4 volunteers for the publication of this report. An outline of the study is summarized in Fig. 1.
FIGURE 1.
Workflow of the study. Five commercial extraction kits were compared for 4 frozen fecal samples.
Bacterial DNA was extracted with 5 commercial DNA extraction kits (Table 1): QIA (Qiagen GmbH), QIAF (Qiagen GmbH), MOB (MO BIO Laboratories), GXT (Hain Lifescience GmbH), and MP (Roche Diagnostics GmbH). Three of the methods (QIA, QIAF, and MOB) are manual extraction methods, of which QIA is 1 of the 2 methods recommended by the IHMS.27 GXT and an identical extraction kit available as DiaSorin Arrow Stool DNA (Fisher Scientific, Pittsburgh, PA, USA), are semiautomatic extraction kits intended to be used with GenoXtract instrument (Hain LifeScience GmbH). MagNA Pure 96 System (Roche Diagnostics GmbH) is a high-throughput, automatic extraction instrument, yet the MP protocol also includes manual preprocessing steps and should thus also be considered as a semiautomatic method. Comparison of the extraction methods included the estimation of the ease of use, DNA gain, and diversity indices and phylogenetic composition identified by the NGS. The suitability of the 2 semiautomated extraction kits GXT or MP in the DNA extraction from stool for NGS analysis has not, in our knowledge, been previously evaluated.
TABLE 1.
DNA extraction methods
| ID | Protocol | Producer | Procedure | Sample weight, mg | Elution volume, µl | Lysis method | DNA capture |
|---|---|---|---|---|---|---|---|
| QIA | QIAamp DNA Stool Mini Kit | Qiagen GmbH | Manual | 200 | 200 | Chemical | Silica membrane |
| QIAF | QIAamp Fast DNA Stool Mini Kit | Qiagen GmbH | Manual | 200 | 200 | Chemical | Silica membrane |
| MOB | PowerFecal DNA Isolation Kit | MO BIO Laboratories | Manual | 100 | 100 | Bead-beating | Silica membrane |
| GXT | GXT Stool Extraction Kit, Ver. 2.0 | Hain Lifescience GmbH | Semiautomatic | 50–100 | 200 | Bead-beating | Magnetic beads |
| MP | MagNA Pure 96 DNA and Viral NA Large Volume Kit | Roche Diagnostics GmbH | Semiautomatic | 50–100 | 100 | Chemical | Magnetic beads |
The 16S rRNA gene profile of the samples was analyzed with 2 different 16S rRNA gene-targeting NGS methods (Fig. 1), 1 targeting the V4–V5 regions of the bacterial 16S rRNA gene and the other targeting the V3–V4 regions of the 16S rRNA gene. The different 16S rDNA sequencing protocols were compared based on the average diversity indices and phylogenetic composition identified by the NGS.
Sample Preprocessing
Stool specimens were homogenized by manual mixing, divided into 10 subsamples that were weighted according to the recommendations of 5 different DNA extraction kits (Table 1), labeled with sample numbers and codes representing the corresponding protocols, and stored at −75°C for 21 d. Two parallel subsamples were weighted for each extraction protocol (Fig. 1).
DNA Extraction
DNA extractions were performed after the 21-d freezer storage of the specimens. Samples were thawed gently on ice, and the DNA was extracted with 5 different protocols (Fig. 1). The extractions were otherwise performed according to the manufacturers’ instructions, but in the semiautomated GXT protocol, the sample vortexing step was replaced with a bead-beating homogenization with MO BIO PowerLyzer 24 Bench Top Bead-Based Homogenizer (MO BIO Laboratories) in 1.4 mm Ceramic Bead Tubes (MO BIO Laboratories) to enhance the cell lysis. The total number of extractions was 40, as 2 parallel extractions were performed with 5 methods for 4 original specimens. The DNA concentrations of the extracts were measured fluorometrically with the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Waltham, MA, USA), after which the DNAs were stored at −75°C until 16S rDNA library preparation.
16S rRNA Gene Sequencing
The microbial 16S rDNA profiles of the 40 DNA extracts were analyzed with 2 distinct MiSeq 16S rRNA gene-sequencing protocols (Illumina). One was an in-house protocol targeting the V4–V5 regions of the bacterial 16S rRNA gene, whereas the other was targeting the V3–V4 regions of the 16S rRNA gene and strictly followed the Illumina 16S Metagenomic Sequencing Library Preparation guide.29 The V4–V5 library preparation and sequencing were performed at the University of Turku (Turku, Finland), whereas the V3–V4 library preparation and sequencing were performed at FISABIO (Valencia, Spain). The V3–V4 region of the bacterial 16S rRNA gene was amplified by following the Illumina 16S Metagenomic Sequencing Library Preparation guide.29 The V4–V5 region was amplified using HiFi PCR kit (KAPA Biosystems, Wilmington, MA, USA) with in-house-generated, indexed primers, modified from Kozich et al.30 The forward and reverse primer sequences for the V4–V5 rRNA gene library preparation are represented in Table 2. The composition of a PCR reaction was as follows: 0.3 μM primers, 0.3 mM dNTPs, 0.5 U polymerase enzyme, and 50 ng DNA template. The PCR program with Veriti Thermal Cycler (Thermo Fisher Scientific) consisted of the following steps: initial denaturation at 98°C for 4 min, 25 cycles at 98°C for 20 s, 65°C for 20 s and 72°C for 35 s, and a final extension at 72°C for 10 min. The expected PCR product size was ∼500 bp. The PCR products were purified with Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA) on the DynaMag-96 Side Magnet (Thermo Fisher Scientific). The PCR products were quality controlled with TapeStation (Agilent Technologies, Santa Clara, CA, USA), and the final DNA concentrations of the purified products were measured with a Qubit 2.0 fluorometer (Thermo Fisher Scientific). The purified products were mixed in equal molar concentrations to generate a 4 nM library pool. The pool was denatured,29 diluted into a final concentration of 4 pM, and spiked with 25% denatured PhiX Control (Illumina) for sequencing. The 16S rRNA gene libraries were sequenced with 2 × 300 bp paired-end reads on the MiSeq systems (Illumina), using MiSeq v3 reagent kits (Illumina). Quality of the raw sequence data was checked with the FastQC quality-control tool (Babraham Bioinformatics, Cambridge, United Kingdom; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the datasets were analyzed with QIIME 1.9 pipeline (Quantitative Insights Into Microbial Ecology; http://qiime.org), as described previously,26, 31, 32 using the GreenGenes 13.08 database.33 Sequence reads were filtered with a quality-score acceptance rate of 20 or better, and the generated operational taxonomic unit (OTU) table was filtered by dropping out OTUs representing <0.05% of the total sequence count. Then, to minimize the effect of intersample variation in the sequencing efficiency, samples were subsampled (rarefied) by random sampling without replacement to the lowest common sequencing depth; i.e., the OTU counts were normalized to match the sample with the lowest total OTU count. This rarefaction level was 8, 766 OTUs in the V3–V4 analysis; 15, 593 OTUs in the V4–V5 analysis; and 16, 741 OTUs in the combined analysis. In preliminary analyses, the results of the duplicates—i.e., the 2 parallel extractions from each sample with each DNA extraction protocol—were found nearly identical. Therefore, for the final result analyses, the read sequences of the duplicates were merged together, forming 1 sample from each 2 replicates. This resulted in a final sample cohort of 20 + 20 samples (Table 3). Three samples from the V3–V4 sequencing run (2.2. GXT, 3.2. MOB, and 4.2. QIA) were discarded as a result of significantly deviating results, but as the quality of the sample duplicates of these samples was good, this had no effect on the final sample number.
TABLE 2.
Primers used for encoding the V4–V5 region of 16S rDNA
| Direction | Primer |
|---|---|
| Forward |
5′-AATGATACGGCGACCACCGAGATCTACAC-i5a-TATGGTAATTGTGTGCCAGCMGCCGCGGTAA-3′ |
| Reverse | 5′-CAAGCAGAAGACGGCATACGAGAT-i7a-AGTCAGTCAGGCCCCGTCAATTCMTTTRAGT-3′ |
i5 and i7 represent 8nt index sequences that enable the identification of sequences originated from each prespecified DNA sample.
TABLE 3.
Final sample cohort
| NGS sample ID | Original sample ID | Extraction method | 16S rRNA gene region |
|---|---|---|---|
| 1033 | 1.1 + 1.2 | QIA | V3–V4 |
| 1035 | 1.1 + 1.2 | QIAF | V3–V4 |
| 1037 | 1.1 + 1.2 | GXT | V3–V4 |
| 1039 | 1.1 + 1.2 | MOB | V3–V4 |
| 1041 | 1.1 + 1.2 | MP | V3–V4 |
| 1043 | 2.1 + 2.2 | QIA | V3–V4 |
| 1045 | 2.1 + 2.2 | QIAF | V3–V4 |
| 47 | 2.1 | GXT | V3–V4 |
| 1049 | 2.1 + 2.2 | MOB | V3–V4 |
| 1051 | 2.1 + 2.2 | MP | V3–V4 |
| 1053 | 3.1 + 3.2 | QIA | V3–V4 |
| 1055 | 3.1 + 3.2 | QIAF | V3–V4 |
| 1057 | 3.1 + 3.2 | GXT | V3–V4 |
| 59 | 3.1 | MOB | V3–V4 |
| 1061 | 3.1 + 3.2 | MP | V3–V4 |
| 63 | 4.1 | QIA | V3–V4 |
| 1065 | 4.1 + 4.2 | QIAF | V3–V4 |
| 1067 | 4.1 + 4.2 | GXT | V3–V4 |
| 1069 | 4.1 + 4.2 | MOB | V3–V4 |
| 1071 | 4.1 + 4.2 | MP | V3–V4 |
| 1133 | 1.1 + 1.2 | QIA | V4–V5 |
| 1135 | 1.1 + 1.2 | QIAF | V4–V5 |
| 1137 | 1.1 + 1.2 | GXT | V4–V5 |
| 1139 | 1.1 + 1.2 | MOB | V4–V5 |
| 1141 | 1.1 + 1.2 | MP | V4–V5 |
| 1143 | 2.1 + 2.2 | QIA | V4–V5 |
| 1145 | 2.1 + 2.2 | QIAF | V4–V5 |
| 1147 | 2.1 + 2.2 | GXT | V4–V5 |
| 1149 | 2.1 + 2.2 | MOB | V4–V5 |
| 1151 | 2.1 + 2.2 | MP | V4–V5 |
| 1153 | 3.1 + 3.2 | QIA | V4–V5 |
| 1155 | 3.1 + 3.2 | QIAF | V4–V5 |
| 1157 | 3.1 + 3.2 | GXT | V4–V5 |
| 1159 | 3.1 + 3.2 | MOB | V4–V5 |
| 1161 | 3.1 + 3.2 | MP | V4–V5 |
| 1163 | 4.1 + 4.2 | QIA | V4–V5 |
| 1165 | 4.1 + 4.2 | QIAF | V4–V5 |
| 1167 | 4.1 + 4.2 | GXT | V4–V5 |
| 1169 | 4.1 + 4.2 | MOB | V4–V5 |
| 1171 | 4.1 + 4.2 | MP | V4–V5 |
Statistical Analyses
To examine the differences in DNA gain among the extraction methods, JMP Pro 12 (SAS Institute, Cary, NC, USA) was used; a nonparametric Kruskal-Wallis test was applied to assess whether any differences occurred, and Steel-Dwass All Pairs test was used to assess the pairwise differences among the methods. P < 0.05 was considered as statistically significant.
Statistical analyses of the 16S rRNA gene sequence data were performed together with QIIME statistical tools and JMP Pro 12. All analyses were made from the randomly subsampled OTU table, with rarefaction level matching the sample with the lowest total OTU count. To study the bacterial diversity of the samples, α-diversity metrics were computed, and α-rarefaction plots were generated with QIIME. Statistically significant differences in the α-diversity (i.e., in average Shannon index values) were then assessed with JMP Pro 12, applying nonparametric methods and considering P < 0.05 as statistically significant.
To test for statistically significant differences in taxonomic richness, i.e., in the OTU abundances, Kruskal-Wallis test was applied with both QIIME and JMP Pro 12. In QIIME, OTUs existing in <25% of the samples were filtered away before testing. Taxonomic levels phylum and genus were studied, and false discovery rate (FDR)-adjusted P < 0.05 was considered as statistically significant. In JMP, the randomly subsampled OTU table was used, and P < 0.05 was considered as statistically significant. If Kruskal-Wallis reported statistical significance for a bacterial phylum/genus between the DNA extraction methods or the study subjects, then Steel-Dwass All Pairs test was used to study the pairwise differences between the groups.
To analyze the differences in the overall bacterial diversity across the samples, weighted UniFrac distance matrices were generated from the randomly subsampled OTU table, and principal coordinate analysis (PCoA) plots were produced. These PCoA plots were visualized with the EMPeror data visualization program. To ascertain whether the visually observed differences were statistically significant, adonis analyses were performed. Adonis returns a P value for significance, alongside an R2 value, indicative of the amount of variation explained by a specific grouping variable (DNA extraction, study subject, or 16S rRNA gene primers).
RESULTS
DNA Extraction Protocol Had Little Influence on the 16S rRNA Gene-Sequencing Results
All evaluated DNA extraction kits produced a decent quantity of DNA from human fecal specimens for NGS analysis (Table 4). However, the average DNA gain (microgram per gram of feces) was significantly higher with the semiautomated GXT than with other methods (Steel-Dwass All Pairs, P < 0.01 for all). In addition, the DNA gain with QIA was higher than with MOB and QIAF (P < 0.01 for both).
TABLE 4.
DNA gain with different extraction protocols
| Sample number | Extraction method | Original sample weight, mg | DNA concentration, ng/µl | DNA gain, µg | DNA gain from gram of original sample, µg/g |
|---|---|---|---|---|---|
| 1.1 | QIA | 198 | 22.4 | 4.5 | 22.6 |
| QIAF | 200 | 6.1 | 1.2 | 6.1 | |
| GXT | 68 | 95.8 | 19.2 | 281.8 | |
| MOB | 76 | 3.2 | 0.3 | 4.2 | |
| MP | 74 | 26.2 | 2.6 | 35.4 | |
| 1.2 | QIA | 202 | 18.0 | 3.6 | 17.8 |
| QIAF | 198 | 2.1 | 0.4 | 2.1 | |
| GXT | 68 | 75.6 | 15.1 | 222.4 | |
| MOB | 89 | 1.8 | 0.2 | 2.0 | |
| MP | 80 | 40.2 | 4.0 | 50.3 | |
| 2.1 | QIA | 200 | 14.4 | 2.9 | 14.4 |
| QIAF | 195 | 1.7 | 0.3 | 1.8 | |
| GXT | 65 | 24.4 | 4.9 | 75.1 | |
| MOB | 103 | 5.3 | 0.5 | 5.1 | |
| MP | 78 | 22.0 | 2.2 | 28.2 | |
| 2.2 | QIA | 195 | 15.8 | 3.2 | 16.2 |
| QIAF | 206 | 3.4 | 0.7 | 3.3 | |
| GXT | 67 | 65.2 | 13.0 | 194.6 | |
| MOB | 104 | 5.7 | 0.6 | 5.5 | |
| MP | 75 | 10.8 | 1.1 | 14.4 | |
| 3.1 | QIA | 188 | 28.3 | 5.7 | 30.1 |
| QIAF | 190 | 6.2 | 1.2 | 6.5 | |
| GXT | 60 | 102.0 | 20.4 | 340.0 | |
| MOB | 103 | 3.6 | 0.4 | 3.5 | |
| MP | 80 | 29.8 | 3.0 | 37.3 | |
| 3.2 | QIA | 200 | 32.1 | 6.4 | 32.1 |
| QIAF | 197 | 8.6 | 1.7 | 8.7 | |
| GXT | 65 | 99.4 | 19.9 | 305.8 | |
| MOB | 105 | 11.1 | 1.1 | 10.6 | |
| MP | 87 | 16.6 | 1.7 | 19.1 | |
| 4.1 | QIA | 175 | 30.7 | 6.1 | 35.1 |
| QIAF | 198 | 4.7 | 0.9 | 4.8 | |
| GXT | 60 | 85.6 | 17.1 | 285.3 | |
| MOB | 96 | 2.6 | 0.3 | 2.7 | |
| MP | 90 | 14.2 | 1.4 | 15.8 | |
| 4.2 | QIA | 170 | 24.5 | 4.9 | 28.8 |
| QIAF | 180 | 3.7 | 0.7 | 4.1 | |
| GXT | 62 | 86.8 | 17.4 | 280.0 | |
| MOB | 95 | 4.7 | 0.5 | 4.9 | |
| MP | 95 | 1.2 | 0.1 | 1.2 |
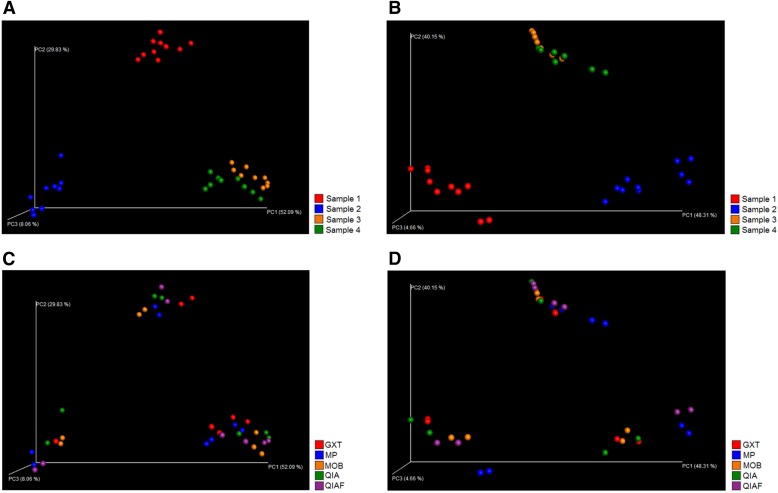
The observed bacterial diversity of the samples, represented as average Shannon index values, was not dependent on the DNA extraction protocol (P = 0.89; Fig. 2B), whereas clear interindividual differences were seen (P < 0.0001; Fig. 2A). The “observed species” metric within QIIME confirmed these findings, presenting little difference among the DNA extraction protocols and notable differences among the study subjects with both V3–V4 sequencing (Fig. 3A and C) and V4–V5 sequencing (Fig. 3B and D). In PCoA, where samples obtaining similar microbiota profiles cluster together, the original specimens 1 and 2 clustered separately with both V3–V4 sequencing (Fig. 4A) and V4–V5 sequencing (Fig. 4B), whereas the original specimens 3 and 4 clustered together (Fig. 4A and B). The subsamples of each original specimen clustered together despite the DNA extraction protocol in V3–V4 sequencing (Fig. 4C). In V4–V5 sequencing, however, minor differences could be observed; subsamples extracted with MP seemed to cluster separately from the other subsamples (Fig. 4D). However, in jack-knifed unweighted pair group method with arithmetic mean (UPGMA) tree (Fig. 5), the subsamples of each original specimen clustered together despite the DNA extraction method, and no statistically significant differences among the extraction methods were observed in the adonis analysis of the microbial community profiles (Table 5). Furthermore, in the phylum-level bacterial composition, i.e., in the OTU abundances, no differences were found in either V3–V4 or V4–V5 sequencing when comparing the samples extracted with different protocols. Interestingly, however, samples processed with MP tended to have a higher Firmicutes-to-Bacteroidetes ratio than samples processed with other DNA extraction methods (P = 0.08). This tendency was not seen in V3–V4 sequencing (P = 0.2). In contrast to the DNA extraction, sample grouping by study subjects was statistically significant in adonis analysis (Table 5). Furthermore, the phylum-level bacterial composition differed significantly among the subjects in both V3–V4 and V4–V5 sequencing (Kruskal-Wallis, P < 0.01 for all).
FIGURE 2.
Bacterial diversity of the samples, represented as average Shannon indices capped with minimum sample size. The average Shannon indices vary significantly among the study subjects (A), whereas no differences are seen among the samples processed with different DNA extraction protocols (B). The average Shannon indices are significantly higher in the V3–V4 samples than in the V4–V5 samples (C).
FIGURE 3.
Bacterial diversity of the samples, represented as observed different OTUs per sequences. No significant differences are inspected in the number of observed OTUs in the samples processed with different DNA extraction protocols in either V3–V4 sequencing (A) or V4–V5 sequencing (B). By contrast, clear interindividual differences can be seen with both methods (C and D). In V3–V4 sequence analysis, samples were rarefied to level 8, 766 reads per sample, whereas in V4–V5 sequence analysis, the rarefaction level was 15, 593 reads per sample.
FIGURE 4.
PCoA (weighted UniFrac) plot. Results of the V3–V4 sequencing are presented on the left (A and C), whereas results of the V4–V5 sequencing are presented on the right (B and D). With both sequencing methods, subsamples of original specimens 3 and 4 cluster together, whereas subsamples of specimens 1 and 2 constitute individual clusters (A and B). In V3–V4 sequencing, the subsamples of each original specimen cluster well together despite the DNA extraction method (C). In V4–V5 sequencing, however, some differences can be seen among the extraction methods (D). In V3–V4 sequence analysis, the rarefaction level was 8, 766 reads per sample, whereas in V4–V5 sequence analysis, the rarefaction level was 15, 593 reads per sample.
FIGURE 5.
Jack-knifed UPGMA tree. When V3–V4 and V4–V5 sequencing results are combined, 2 clearly separate clusters are formed. Inside these clusters, subsamples of each original specimen cluster together despite the DNA extraction method. The rarefaction level was 16, 741 reads per sample.
TABLE 5.
Adonis test results
| Extraction method |
Study subject |
16S rDNA primers |
||||
|---|---|---|---|---|---|---|
| R2 | P | R2 | P | R2 | P | |
| V3–V4 sequence analysis | 0.05101 | 0.972 | 0.42132a | 0.001a | – | – |
| V4–V5 sequence analysis | 0.06062 | 0.882 | 0.44237a | 0.001a | – | – |
| Combined analysis | 0.01611 | 1.000 | 0.16903 | 0.001b | 0.51328 | 0.001b |
The low P values indicate that the grouping of samples by study subjects is statistically significant in both V3–V4 and V4–V5 sequencing. The R2 values indicate that ∼43% of the variation in distances can be explained by this grouping.
The low P values indicate that the sample grouping by study subjects and by 16S rDNA primers are statistically significant. The R2 values indicate that ∼17% and 51% of the variation in distances can be explained by these groupings, respectively.
In the bacterial genus level, statistically significant differences could be seen among the DNA extraction methods in the genera Dorea and Coprococcus (Kruskal-Wallis FDR adjusted, P < 0.05): in V3–V4 sequencing, Dorea was found more abundant with MP than with other methods (Steel-Dwass, P < 0.05 for all) and more abundant with MOB than with QIA and QIAF (P < 0.05 for both), whereas Coprococcus was found more abundant with GXT than with MP and QIA (P < 0.05 for both). In V4–V5 sequencing, Dorea was found more abundant with MP than with QIA and QIAF (P < 0.05 for both) and more abundant with MOB than with QIA (P < 0.05), whereas Coprococcus was found more abundant with QIAF, GXT, and MOB than with QIA (P < 0.05 for all). No other statistically significant differences were found with either QIIME or JMP Pro 12 between the samples extracted with different DNA extraction protocols. Intrasubject variation, by contrast, was observed in the vast majority of the bacterial genera (results not shown).
16S rDNA Primers Had a Substantial Effect on the 16S rRNA Gene-Sequencing Results
The 16S rRNA gene profiles produced by the 2 sequencing protocols differed considerably from each other. The number of recognized OTUs was significantly higher in the V4–V5 samples than in the V3–V4 samples (Mann-Whitney U test, P < 0.001). Nevertheless, the observed microbial diversity, represented as the average Shannon index values, capped with the lowest total OTU count, was significantly higher in the V3–V4 samples (P < 0.001; Fig. 2C and 6). In the PCoA plot, the samples analyzed with the different sequencing protocols clustered separately (Fig. 7), and in addition, 2 clearly separate clusters were formed in the jack-knifed UPGMA tree (Fig. 5). These observations were supported by the adonis analysis rarefied to 16, 741 sequences per sample for weighted UniFrac; based on adonis, a remarkable portion of variation between the samples could be attributed to the 16S rDNA primers (Table 5).
FIGURE 6.
Bacterial diversity of the samples analyzed with different 16S rRNA gene-sequencing methods, represented as observed different OTUs per sequences. The average diversity is significantly higher in the samples analyzed with V3–V4 sequencing (P < 0.001).
FIGURE 7.
PCoA (weighted UniFrac) plot of all analyzed samples. When V3–V4 and V4–V5 sequencing results are combined, each original specimen comprises 2 separate clusters (A), as the subsamples analyzed with different 16S rDNA sequencing methods clearly cluster separately (B). Rarefaction level was 16, 741 reads per sample.
In the bacterial phylum level, QIIME reported statistically significant differences between the V3–V4 and V4–V5 sequencing in the abundance of the phyla Lentisphaerae, Actinobacteria, Bacteroidetes, Proteobacteria, and Firmicutes (FDR, P < 0.05 for all); the average Lentisphaerae and Bacteroidetes abundances were higher with V4–V5 sequencing, whereas the abundances of the other phyla were higher with V3–V4 sequencing. However, the difference in Lentisphaerae was not statistically significant when analyzed with JMP Pro 12 (P = 0.59). The Firmicutes-to-Bacteroidetes ratio was significantly lower in V4–V5 sequencing (P < 0.01).
In the bacterial genus level, QIIME reported statistically significant differences in 21 genera between the V3–V4 and V4–V5 sequencing protocols. For example, the genus Parabacteroides was significantly more abundant in the samples analyzed with V4–V5 sequencing (FDR, P < 0.05), whereas Bifidobacterium, Coprococcus, and Blautia were more abundant in the V3–V4 samples (FDR, P < 0.05 for all). In addition, the genera Sphingomonas, Roseburia, and Bilophila were detectable only with V3–V4 sequencing, whereas Clostridium and Lactococcus could only be detected with V4–V5 sequencing.
DISCUSSION
The rapid progression of the NGS methods has revolutionized the microbiota research, as the high-throughput protocols have become more cost effective and thus, more readily available.8, 9 Still to date, surprisingly, little attention has been drawn on the general quality control of the NGS-based approaches used in the growing field of human microbiota research, even though it is a well-known fact that analysis methods in this study field are highly sensitive and thus, prone to biases. To gain a deeper understanding of the possible bias-generating steps in the NGS procedures and to enhance data quality and result comparability, we analyzed both the impact of different DNA extraction methods and 16S rRNA gene-targeting primers on the gut microbiota profiles. Special attention in this study was paid to the evaluation of the suitability of 2 commercially available semiautomated DNA extraction methods for 16S rRNA gene sequencing, as DNA extraction is often the bottleneck of otherwise high-throughput NGS studies. Based on our results, the chosen targeted 16S rRNA gene region has a major impact on the gut microbiota profiles. This finding is in line with several previous studies.20–23, 25 Furthermore, our results suggest that the semiautomated DNA extraction methods are efficient, feasible, and suitable for the gut microbiota NGS applications. As a matter of fact, our results propose that the overall effect of DNA extraction on the 16S rRNA gene-sequencing results is relatively small.
All commercial DNA extraction kits evaluated in this study showed sufficient performance in extracting bacterial DNA from human feces. However, the DNA gain was higher with GXT compared with other methods. This can be partly explained by the extra mechanical lysis step added to the GXT protocol, as bead-beating has been previously shown to enhance the DNA yield from fecal samples.19, 34 In addition, some previous reports suggest that spin column-based DNA extraction methods have a limited DNA-binding capacity.17 As MOB also includes a bead-beating step, and MP is not based on the spin columns, the variation in the extraction efficiency is likely to result from differences in the extraction chemistries. However, as the 16S rDNA library preparation was successful regardless of the extraction kit, it can be concluded that the DNA quantity and quality were sufficient with all tested methods. The previously reported gut microbiota profile differences, depending on the manual extraction methods,19, 35 may, at least to some extent, arise from the human errors, such as pipetting. Thus, the desirable results gained from the semiautomated methods are of practical importance. In addition, with the consideration of the amount of labor affiliated with each extraction protocol, GXT and MP obviously rise above the manual methods. GXT can extract DNA from 1 to 12 samples in a single, 1 h run, and the method is thus suitable for daily use in various study setups. By contrast, MP uses 96-well plates and is best suited for bulk extractions of large sample sets. However, the DNA gain with MP was ∼8 times lower than with GXT, and therefore, the extraction of a sufficient amount of DNA from very scarce samples may be challenging with MP.
Several previous studies have reported notable variation among different DNA extraction methods.17–19, 35–37 However, no clear consensus on the subject has been reached, as each study has applied different methodologies, and the observed differences vary among the studies. Interestingly, in this study, only minor differences were observed among the extraction methods. The samples extracted with different protocols clustered together in the PCoA, and no variation in the average Shannon index values was observed. In V4–V5 sequencing, however, samples extracted with semiautomatic MP method tended to have higher Firmicutes-to-Bacteroidetes abundance, suggesting that slight differences in the capability to extract DNA from Gram-positive bacteria occurred between the protocols. This finding is in line with previous studies that have reported notable differences among DNA extraction methods in the ability to isolate DNA of Gram-positive bacteria.35, 38 Regarding the Gram-positive bacteria, 1 especially troublesome genus has been Bifidobacterium; it has been reported that Bifidobacterium cannot be adequately detected without an effective bead-beating step in the DNA extraction.38 In this study, no differences in the Bifidobacterium abundance were seen among the DNA extraction protocols, with or without bead-beating. However, the observed Bifidobacterium abundance was extremely low with all of the DNA extraction methods used, suggesting that the low Bifidobacterium abundance in this study results either from the 16S rDNA library preparation protocols or sample material. As Bifidobacterium abundance has been shown to increase in, e.g., diet interventions and further associate with the loss of weight or fat mass,39, 40 detection of Bifidobacterium may be important in interventional human studies that aim at weight reduction. However, despite that Bifidobacterium may represent up to 14% of human gut microbiota,41 significantly lower abundances have been reported in several studies.41, 42 In light of these controversies, emphasis should be put on better characterization of Bifidobacterium among human gut microbiota.
As previously reported20, 22, 23, 25 and reaffirmed in this study, the selected target variable region of the 16S rRNA gene has a major impact on the analysis results. The samples amplified with different primers clustered clearly separately in PCoA, and significant differences in the bacterial composition were observed in both phylum and genus level. Furthermore, the overall bacterial diversity, represented as average Shannon index values, was significantly higher in the samples analyzed with the V3–V4 protocol. In addition, both methods failed to detect certain bacterial genera that were observed with the other method. This may have a significant impact on the interpretation of the data, for instance, regarding the commonly used parameter, Firmicutes-to-Bacteroidetes ratio. In fact, the inconsistencies in the association between Firmicutes abundance and obesity43–46 may, at least partly, arise from the different analysis techniques applied in the studies.
Overall, the reliable interpretation of microbiota NGS data is hampered by various sources of uncertainty. In addition to raw sequence quality, the intactness, as well as reliability, of the sequencing results is dependent on the selected gene database and data-filtering protocols.9 In this study, previously described quality-filtering protocols were used to enhance the result reliability.26, 32 In addition, before the statistical analyses with QIIME, the OTU count of each sample was normalized to match the sample with the lowest total OTU count. The purpose of this normalization was to minimize the possible bias caused by the variation in the sequencing efficiency between the samples. One solution for enhancing the reliability of the NGS protocol could be the use of 2 or more primer sets simultaneously to cover the bacterial diversity in complex samples more comprehensively. We hypothesize that this could be achieved by using a combination of primers differing from each other in key positions defining bacterial strain specificity. By using this type of approach, it could be possible to analyze bacteria that are traditionally hard to capture with a single primer-pair approach. Before applying this approach on actual studies, the different primer sequences and combinations should be thoroughly tested by using well-characterized control samples containing known bacterial strains in well-quantified proportions.
This study was limited by the fact that only the 16S rRNA gene was sequenced instead of the genomic sequencing of the whole microbiome. It is possible that variable copurification of eukaryotic DNA (human, fungal, food, or plant origin) with different DNA extraction methods has a more prominent effect on the whole-genome sequencing results than on the results from 16S rRNA gene-targeting-based methods. DNA extraction has recently been reported to influence the fecal microbiota community structure when sequencing the whole gut microbiome with the Illumina HiSeq system, but even then, the interindividual variation in the samples clearly exceeded the variation resulting from choice of extraction method.17 Nevertheless, the results gained in this study are applicable only for the bacterial composition analyses, whereas further studies are required to reveal the effect of the DNA extraction on the functional microbiome analyses and whole-genome sequencing results.
CONCLUSIONS
The results of this study suggest that semiautomatic DNA extraction protocols offer a practical and functional option for the manual methods in the preprocessing of fecal samples for gut microbiota composition analyses. Automation could reduce variability between experiments and enhance the sample preprocessing steps, thus enabling more high-throughput study setups in the gut microbiota research field. These methods could also be easily applied to routine diagnostic settings in clinical laboratories. Furthermore, this study shows that the 16S rRNA gene target region has a major impact on the gut microbiota NGS results. This underlines the importance of careful selection of the 16S rRNA gene-targeting primers and emphasizes that extreme caution should be taken when comparing studies conducted with different 16S rRNA gene-sequencing methods.
ACKNOWLEDGMENTS
The authors warmly thank Heidi Isokääntä, Katri Kylä-Mattila, and Jukka Karhu for their valuable technical assistance in this study. A.R. received financial support from the Turku University Foundation. S. Pekkala is an Academy of Finland postdoctoral fellow.
REFERENCES
- 1.Eckburg PB, Bik EM, Bernstein CN, et al. . Diversity of the human intestinal microbial flora. Science 2005;308:1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev 2010;90:859–904. [DOI] [PubMed] [Google Scholar]
- 3.Bäckhed F, Ding H, Wang T, et al. . The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004;101:15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker C, Neurath MF, Wirtz S. The intestinal microbiota in inflammatory bowel disease. ILAR J 2015;56:192–204. [DOI] [PubMed] [Google Scholar]
- 5.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006;444:1022–1023. [DOI] [PubMed] [Google Scholar]
- 6.Walker AW, Lawley TD. Therapeutic modulation of intestinal dysbiosis. Pharmacol Res 2013;69:75–86. [DOI] [PubMed] [Google Scholar]
- 7.Guinane CM, Cotter PD. Role of the gut microbiota in health and chronic gastrointestinal disease: understanding a hidden metabolic organ. Therap Adv Gastroenterol 2013;6:295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tringe SG, Hugenholtz P. A renaissance for the pioneering 16S rRNA gene. Curr Opin Microbiol 2008;11:442–446. [DOI] [PubMed] [Google Scholar]
- 9.Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 2009;19:1141–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apajalahti JH, Kettunen A, Nurminen PH, Jatila H, Holben WE. Selective plating underestimates abundance and shows differential recovery of bifidobacterial species from human feces. Appl Environ Microbiol 2003;69:5731–5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tannock GW. Analysis of the intestinal microflora using molecular methods. Eur J Clin Nutr 2002;56 (Suppl 4): S44–S49. [DOI] [PubMed] [Google Scholar]
- 12.Zoetendal EG, Vaughan EE, de Vos WM. A microbial world within us. Mol Microbiol 2006;59:1639–1650. [DOI] [PubMed] [Google Scholar]
- 13.Qin J, Li R, Raes J, et al. . A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso JG, Lauber CL, Walters WA, et al. . Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandal RS, Saha S, Das S. Metagenomic surveys of gut microbiota. Genomics Proteomics Bioinformatics 2015;13:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 2009;326:1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wesolowska-Andersen A, Bahl MI, Carvalho V, et al. . Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2014;2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burbach K, Seifert J, Pieper DH, Camarinha‐Silva A. Evaluation of DNA extraction kits and phylogenetic diversity of the porcine gastrointestinal tract based on Illumina sequencing of two hypervariable regions. Microbiologyopen 2016;5:70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maukonen J, Simões C, Saarela M. The currently used commercial DNA-extraction methods give different results of clostridial and actinobacterial populations derived from human fecal samples. FEMS Microbiol Ecol 2012;79:697–708. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 2008;36:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 2007;73:5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klindworth A, Pruesse E, Schweer T, et al. . Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Starke IC, Vahjen W, Pieper R, Zentek J. The influence of DNA extraction procedure and primer set on the bacterial community analysis by pyrosequencing of barcoded 16S rRNA gene amplicons. Mol Biol Int 2014;2014:548683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD. 16S rRNA gene sequencing of mock microbial populations—impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol 2016;16:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremblay J, Singh K, Fern A, et al. . Primer and platform effects on 16S rRNA tag sequencing. Front Microbiol 2015;6:771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bokulich NA, Subramanian S, Faith JJ, et al. . Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 2013;10:57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.International Human Microbiome Standards. Updated 2015. Retrieved July 15, 2016, from http://www.microbiome-standards.org.
- 28.Claassen S, du Toit E, Kaba M, Moodley C, Zar HJ, Nicol MP. A comparison of the efficiency of five different commercial DNA extraction kits for extraction of DNA from faecal samples. J Microbiol Methods 2013;94:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.16S Metagenomic Sequencing Library Preparation San Diego, CA: Illumina. Updated 2014. Retrieved July 15, 2016, from https://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf.
- 30.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013;79:5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol 2012:Chapter 1:Unit 1E.5. [DOI] [PMC free article] [PubMed]
- 33.DeSantis TZ, Hugenholtz P, Larsen N, et al. . Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72:5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariefdjohan MW, Savaiano DA, Nakatsu CH. Comparison of DNA extraction kits for PCR-DGGE analysis of human intestinal microbial communities from fecal specimens. Nutr J 2010;9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner Mackenzie B, Waite DW, Taylor MW. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Front Microbiol 2015;6:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015;3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy NA, Walker AW, Berry SH, et al. . The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One 2014;9:e88982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 2015;3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dewulf EM, Cani PD, Claus SP, et al. . Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013;62:1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao S, Fei N, Pang X, et al. . A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS Microbiol Ecol 2014;87:357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arboleya S, Watkins C, Stanton C, Ross RP. Gut bifidobacteria populations in human health and aging. Front Microbiol 2016;7:1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yatsunenko T, Rey FE, Manary MJ, et al. . Human gut microbiome viewed across age and geography. Nature 2012;486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005;102:11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jumpertz R, Le DS, Turnbaugh PJ, et al. . Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr 2011;94:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pekkala S, Munukka E, Kong L, et al. . Toll-like receptor 5 in obesity: the role of gut microbiota and adipose tissue inflammation. Obesity (Silver Spring) 2015;23:581–590. [DOI] [PubMed] [Google Scholar]
- 46.Liou AP, Paziuk M, Luevano JM Jr, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 2013;5:178ra41. [DOI] [PMC free article] [PubMed] [Google Scholar]