Cyclooxygenase-2
Cyclooxygenase-2 (COX-2), also known as prostaglandin-endoperoxide synthase 2 (HUGO PTGS2), is an enzyme that in humans is encoded by the PTGS2 gene.[5] In humans it is one of three cyclooxygenases. It is involved in the conversion of arachidonic acid to prostaglandin H2, an important precursor of prostacyclin, which is expressed in inflammation.
Function
[edit]PTGS2 (COX-2), converts arachidonic acid (AA) to prostaglandin endoperoxide H2. PTGSs are targets for NSAIDs and PTGS2 (COX-2) specific inhibitors called coxibs. PTGS-2 is a sequence homodimer. Each monomer of the enzyme has a peroxidase and a PTGS (COX) active site. The PTGS (COX) enzymes catalyze the conversion of AA to prostaglandins in two steps. First, hydrogen is abstracted from carbon 13 of arachidonic acid, and then two molecules of oxygen are added by the PTGS2 (COX-2), giving PGG2. Second, PGG2 is reduced to PGH2 in the peroxidase active site. The synthesized PGH2 is converted to prostaglandins (PGD2, PGE2, PGF2α), prostacyclin (PGI2), or thromboxane A2 by tissue-specific isomerases (Figure 2).[6]
While metabolizing arachidonic acid primarily to PGG2, COX-2 also converts this fatty acid to small amounts of a racemic mixture of 15-hydroxyicosatetraenoic acids (i.e., 15-HETEs) composed of ~22% 15(R)-HETE and ~78% 15(S)-HETE stereoisomers as well as a small amount of 11(R)-HETE.[7] The two 15-HETE stereoisomers have intrinsic biological activities but, perhaps more importantly, can be further metabolized to a major class of agents, the lipoxins. Furthermore, aspirin-treated COX-2 metabolizes arachidonic acid almost exclusively to 15(R)-HETE which product can be further metabolized to epi-lipoxins.[8] The lipoxins and epi-lipoxins are potent anti-inflammatory agents and may contribute to the overall activities of the two COX's as well as to aspirin.[citation needed]
COX-2 is naturally inhibited by calcitriol (the active form of vitamin D).[9][10]
Mechanism
[edit]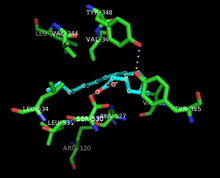

Both the peroxidase and PTGS activities are inactivated during catalysis by mechanism-based, first-order processes, which means that PGHS-2 peroxidase or PTGS activities fall to zero within 1–2 minutes, even in the presence of sufficient substrates.[12][13][14]
The conversion of arachidonic acid to PGG2 can be shown as a series of radical reactions analogous to polyunsaturated fatty acid autoxidation.[15] The 13-pro(S) -hydrogen is abstracted and dioxygen traps the pentadienyl radical at carbon 11. The 11-peroxyl radical cyclizes at carbon 9 and the carbon-centered radical generated at C-8 cyclizes at carbon 12, generating the endoperoxide. The allylic radical generated is trapped by dioxygen at carbon 15 to form the 15-(S) -peroxyl radical; this radical is then reduced to PGG2. This is supported by the following evidence: 1) a significant kinetic isotope effect is observed for the abstraction of the 13-pro (S)-hydrogen; 2) carbon-centered radicals are trapped during catalysis;[16] 3) small amounts of oxidation products are formed due to the oxygen trapping of an allylic radical intermediate at positions 13 and 15.[17][18]
Another mechanism in which the 13-pro (S)-hydrogen is deprotonated and the carbanion is oxidized to a radical is theoretically possible. However, oxygenation of 10,10-difluoroarachidonic acid to 11-(S)-hydroxyeicosa-5,8,12,14-tetraenoic acid is not consistent with the generation of a carbanion intermediate because it would eliminate fluoride to form a conjugated diene.[19] The absence of endoperoxide-containing products derived from 10,10-difluoroarachidonic acid has been thought to indicate the importance of a C-10 carbocation in PGG2 synthesis.[20] However, the cationic mechanism requires that endoperoxide formation comes before the removal of the 13-pro (S)-hydrogen. This is not consistent with the results of the isotope experiments of arachidonic acid oxygenation.[21]
Structure
[edit]
PTGS2 (COX-2) exists as a homodimer, each monomer with a molecular mass of about 70 kDa. The tertiary and quaternary structures of PTGS1 (COX-1) and PTGS2 (COX-2) enzymes are almost identical. Each subunit has three different structural domains: a short N-terminal epidermal growth factor (EGF) domain; an α-helical membrane-binding moiety; and a C-terminal catalytic domain. PTGS (COX, which can be confused with "cytochrome oxidase") enzymes are monotopic membrane proteins; the membrane-binding domain consists of a series of amphipathic α helices with several hydrophobic amino acids exposed to a membrane monolayer. PTGS1 (COX-1) and PTGS2 (COX-2) are bifunctional enzymes that carry out two consecutive chemical reactions in spatially distinct but mechanistically coupled active sites. Both the cyclooxygenase and the peroxidase active sites are located in the catalytic domain, which accounts for approximately 80% of the protein. The catalytic domain is homologous to mammalian peroxidases such as myeloperoxidase.[23][24]
It has been found that human PTGS2 (COX-2) functions as a conformational heterodimer having a catalytic monomer (E-cat) and an allosteric monomer (E-allo). Heme binds only to the peroxidase site of E-cat while substrates, as well as certain inhibitors (e.g. celecoxib), bind the COX site of E-cat. E-cat is regulated by E-allo in a way dependent on what ligand is bound to E-allo. Substrate and non-substrate fatty acids (FAs) and some PTGS (COX) inhibitors (e.g. naproxen) preferentially bind to the PTGS (COX) site of E-allo. Arachidonic acid can bind to E-cat and E-allo, but the affinity of AA for E-allo is 25 times that for Ecat. Palmitic acid, an efficacious stimulator of huPGHS-2, binds only E-allo in palmitic acid/murine PGHS-2 co-crystals. Non-substrate FAs can potentiate or attenuate PTGS (COX) inhibitors depending on the fatty acid and whether the inhibitor binds E-cat or E-allo. Studies suggest that the concentration and composition of the free fatty acid pool in the environment in which PGHS-2 functions in cells, also referred to as the FA tone, is a key factor regulating the activity of PGHS-2 and its response to PTGS (COX) inhibitors.[22]
Clinical significance
[edit]
PTGS2 (COX-2) is unexpressed under normal conditions in most cells, but elevated levels are found during inflammation. PTGS1 (COX-1) is constitutively expressed in many tissues and is the predominant form in gastric mucosa and in the kidneys. Inhibition of PTGS1 (COX-1) reduces the basal production of cytoprotective PGE2 and PG12 in the stomach, which may contribute to gastric ulceration. Since PTGS2 (COX-2) is generally expressed only in cells where prostaglandins are upregulated (e.g., during inflammation), drug-candidates that selectively inhibit PTGS2 (COX-2) were suspected to show fewer side-effects[24] but proved to substantially increase risk for cardiovascular events such as heart attack and stroke. Two different mechanisms may explain contradictory effects. Low-dose aspirin protects against heart attacks and strokes by blocking PTGS1 (COX-1) from forming a prostaglandin called thromboxane A2. It sticks platelets together and promotes clotting; inhibiting this helps prevent heart disease. On the other hand, PTGS2 (COX-2) is a more important source of prostaglandins, particularly prostacyclin which is found in blood vessel lining. Prostacyclin relaxes or unsticks platelets, so selective COX-2 inhibitors (coxibs) increase risk of cardiovascular events due to clotting.[26]
Non-steroidal anti-inflammatory drugs (NSAIDs) inhibit prostaglandin production by PTGS1 (COX-1) and PTGS2 (COX-2). NSAIDs selective for inhibition of PTGS2 (COX-2) are less likely than traditional drugs to cause gastrointestinal adverse effects, but could cause cardiovascular events, such as heart failure, myocardial infarction, and stroke. Studies with human pharmacology and genetics, genetically manipulated rodents, and other animal models and randomized trials indicate that this is due to suppression of PTGS2 (COX-2)-dependent cardioprotective prostaglandins, prostacyclin in particular.[27]
The expression of PTGS2 (COX-2) is upregulated in many cancers. The overexpression of PTGS2 (COX-2) along with increased angiogenesis and SLC2A1 (GLUT-1) expression is significantly associated with gallbladder carcinomas.[28] Furthermore, the product of PTGS2 (COX-2), PGH2 is converted by prostaglandin E2 synthase into PGE2, which in turn can stimulate cancer progression. Consequently, inhibiting PTGS2 (COX-2) may have benefit in the prevention and treatment of these types of cancer.[29][30]
COX-2 expression was found in human idiopathic epiretinal membranes.[31] Cyclooxygenases blocking by lornoxicam in acute stage of inflammation reduced the frequency of membrane formation by 43% in the dispase model of PVR and by 31% in the concanavalin one. Lornoxicam not only normalized the expression of cyclooxygenases in both models of PVR, but also neutralized the changes of the retina and the choroid thickness caused by the injection of pro-inflammatory agents. These facts underline the importance of cyclooxygenases and prostaglandins in the development of PVR.[32]
PTGS2 gene upregulation has also been linked with multiple stages of human reproduction. Presence of gene is found in the chorionic plate, in the amnion epithelium, syncytiotrophoblasts, villous fibroblasts, chorionic trophoblasts, amniotic trophoblasts, as well as the basal plate of the placenta, in the decidual cells and extravillous cytotrophoblasts. During the process of chorioamnionitis/deciduitis, the upregulation of PTGS2 in the amnion and choriodecidua is one of three limited effects of inflammation in the uterus. Increased expression of the PTGS2 gene in the fetal membranes is connected to the presence of inflammation, causing uterine prostaglandin gene expression and immunolocalization of prostaglandin pathway proteins in chorionic trophoblast cells and adjacent decidua, or choriodecidua. PTGS2 is linked with the inflammatory system and has been observed in inflammatory leukocytes. It has been noted that there is a positive correlation with PTGS2 expression in the amnion during spontaneous labour and was discovered to have increased expression with gestational age following the presence of labour with no change observed in amnion and choriodecidua during either preterm or term labour. Additionally, oxytocin stimulates the expression of PTGS2 in myometrial cells.[33]
The mutant allele PTGS2 5939C carriers among the Han Chinese population have been shown to have a higher risk of gastric cancer. In addition, a connection was found between Helicobacter pylori infection and the presence of the 5939C allele.[34]
Interactions
[edit]PTGS2 has been shown to interact with caveolin 1.[35]
History
[edit]PTGS2 (COX-2) was discovered in 1991 by the Daniel Simmons laboratory[36][better source needed] at Brigham Young University.
See also
[edit]- Arachidonic acid
- Cyclooxygenase
- Cyclooxygenase 1
- NSAID
- Discovery and development of COX-2 selective inhibitors
- COX-2 selective inhibitor
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000073756 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000032487 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Hla T, Neilson K (August 1992). "Human cyclooxygenase-2 cDNA". Proc. Natl. Acad. Sci. U.S.A. 89 (16): 7384–8. Bibcode:1992PNAS...89.7384H. doi:10.1073/pnas.89.16.7384. PMC 49714. PMID 1380156.
- ^ O'Banion MK (1999). "Cyclooxygenase-2: molecular biology, pharmacology, and neurobiology". Crit Rev Neurobiol. 13 (1): 45–82. doi:10.1615/critrevneurobiol.v13.i1.30. PMID 10223523.
- ^ Mulugeta S, Suzuki T, Hernandez NT, Griesser M, Boeglin WE, Schneider C (2010). "Identification and absolute configuration of dihydroxy-arachidonic acids formed by oxygenation of 5S-HETE by native and aspirin-acetylated COX-2". J. Lipid Res. 51 (3): 575–85. doi:10.1194/jlr.M001719. PMC 2817587. PMID 19752399.
- ^ Serhan CN (2005). "Lipoxins and aspirin-triggered 15-epi-lipoxins are the first lipid mediators of endogenous anti-inflammation and resolution". Prostaglandins Leukot. Essent. Fatty Acids. 73 (3–4): 141–62. doi:10.1016/j.plefa.2005.05.002. PMID 16005201.
- ^ Wang Q, He Y, Shen Y, Zhang Q, Chen D, Zuo C, Qin J, Wang H, Wang J, Yu Y (2014). "Vitamin D inhibits COX-2 expression and inflammatory response by targeting thioesterase superfamily member 4". J Biol Chem. 289 (17): 11681–11694. doi:10.1074/jbc.M113.517581. PMC 4002078. PMID 24619416.
- ^ Kassi E, Adamopoulos C, Basdra EK, Papavassiliou AG (2013). "Role of Vitamin D in Atherosclerosis". Circulation. 128 (23): 2517–2531. doi:10.1161/CIRCULATIONAHA.113.002654. PMID 24297817.
- ^ PDB: 3OLT
- ^ Smith WL, Garavito RM, DeWitt DL (December 1996). "Prostaglandin endoperoxide H synthases (cyclooxygenases)-1 and -2". J. Biol. Chem. 271 (52): 33157–60. doi:10.1074/jbc.271.52.33157. PMID 8969167.
- ^ Wu G, Wei C, Kulmacz RJ, Osawa Y, Tsai AL (April 1999). "A mechanistic study of self-inactivation of the peroxidase activity in prostaglandin H synthase-1". J. Biol. Chem. 274 (14): 9231–7. doi:10.1074/jbc.274.14.9231. PMID 10092596.
- ^ Callan OH, So OY, Swinney DC (February 1996). "The kinetic factors that determine the affinity and selectivity for slow binding inhibition of human prostaglandin H synthase 1 and 2 by indomethacin and flurbiprofen". J. Biol. Chem. 271 (7): 3548–54. doi:10.1074/jbc.271.7.3548. PMID 8631960.
- ^ Porter NA (1986). "Mechanisms for the autoxidation of polyunsaturated lipids". Accounts of Chemical Research. 19 (9): 262–8. doi:10.1021/ar00129a001.
- ^ Mason RP, Kalyanaraman B, Tainer BE, Eling TE (June 1980). "A carbon-centered free radical intermediate in the prostaglandin synthetase oxidation of arachidonic acid. Spin trapping and oxygen uptake studies". J. Biol. Chem. 255 (11): 5019–22. doi:10.1016/S0021-9258(19)70741-8. PMID 6246094.
- ^ Hecker M, Ullrich V, Fischer C, Meese CO (November 1987). "Identification of novel arachidonic acid metabolites formed by prostaglandin H synthase". Eur. J. Biochem. 169 (1): 113–23. doi:10.1111/j.1432-1033.1987.tb13587.x. PMID 3119336.
- ^ Xiao G, Tsai AL, Palmer G, Boyar WC, Marshall PJ, Kulmacz RJ (February 1997). "Analysis of hydroperoxide-induced tyrosyl radicals and lipoxygenase activity in aspirin-treated human prostaglandin H synthase-2". Biochemistry. 36 (7): 1836–45. doi:10.1021/bi962476u. PMID 9048568.
- ^ Kwok PY, Muellner FW, Fried J (June 1987). "Enzymatic conversions of 10,10-difluoroarachidonic acid with PGH synthase and soybean lipoxygenase". Journal of the American Chemical Society. 109 (12): 3692–3698. doi:10.1021/ja00246a028.
- ^ Dean AM, Dean FM (May 1999). "Carbocations in the synthesis of prostaglandins by the cyclooxygenase of PGH synthase? A radical departure!". Protein Sci. 8 (5): 1087–98. doi:10.1110/ps.8.5.1087. PMC 2144324. PMID 10338019.
- ^ Hamberg M, Samuelsson B (November 1967). "On the mechanism of the biosynthesis of prostaglandins E-1 and F-1-alpha". J. Biol. Chem. 242 (22): 5336–43. doi:10.1016/S0021-9258(18)99433-0. PMID 6070851.
- ^ a b Dong L, Vecchio AJ, Sharma NP, Jurban BJ, Malkowski MG, Smith WL (May 2011). "Human cyclooxygenase-2 is a sequence homodimer that functions as a conformational heterodimer". J. Biol. Chem. 286 (21): 19035–46. doi:10.1074/jbc.M111.231969. PMC 3099718. PMID 21467029.
- ^ Picot D, Loll PJ, Garavito RM (January 1994). "The X-ray crystal structure of the membrane protein prostaglandin H2 synthase-1". Nature. 367 (6460): 243–9. Bibcode:1994Natur.367..243P. doi:10.1038/367243a0. PMID 8121489. S2CID 4340064.
- ^ a b Kurumbail RG, Kiefer JR, Marnett LJ (December 2001). "Cyclooxygenase enzymes: catalysis and inhibition". Curr. Opin. Struct. Biol. 11 (6): 752–60. doi:10.1016/S0959-440X(01)00277-9. PMID 11751058.
- ^ PDB: 3PGH
- ^ Ruan CH, So SP, Ruan KH (2011). "Inducible COX-2 dominates over COX-1 in prostacyclin biosynthesis: Mechanisms of COX-2 inhibitor risk to heart disease". Life Sciences. 88 (1–2): 24–30. doi:10.1016/j.lfs.2010.10.017. PMC 3046773. PMID 21035466.
- ^ Wang D, Patel VV, Ricciotti E, Zhou R, Levin MD, Gao E, Yu Z, Ferrari VA, Lu MM, Xu J, Zhang H, Hui Y, Cheng Y, Petrenko N, Yu Y, FitzGerald GA (May 2009). "Cardiomyocyte cyclooxygenase-2 influences cardiac rhythm and function". Proc. Natl. Acad. Sci. U.S.A. 106 (18): 7548–52. Bibcode:2009PNAS..106.7548W. doi:10.1073/pnas.0805806106. PMC 2670242. PMID 19376970.
- ^ Legan M (August 2010). "Cyclooxygenase-2, p53 and glucose transporter-1 as predictors of malignancy in the development of gallbladder carcinomas". Bosn J Basic Med Sci. 10 (3): 192–6. doi:10.17305/bjbms.2010.2684. PMC 5504494. PMID 20846124.
- ^ EntrezGene 5743
- ^ Menter DG, Schilsky RL, DuBois RN (March 2010). "Cyclooxygenase-2 and cancer treatment: understanding the risk should be worth the reward". Clin. Cancer Res. 16 (5): 1384–90. doi:10.1158/1078-0432.CCR-09-0788. PMC 4307592. PMID 20179228.
- ^ KASE S, SAITO W, OHNO S, ISHIDA S (2010). "Cyclo-Oxygenase-2 Expression in Human Idiopathic Epiretinal Membrane". Retina. 30 (5): 719–723. doi:10.1097/iae.0b013e3181c59698. PMID 19996819. S2CID 205650971.
- ^ Tikhonovich MV, Erdiakov AK, Gavrilova SA (2017-06-21). "Nonsteroid anti-inflammatory therapy suppresses the development of proliferative vitreoretinopathy more effectively than a steroid one". International Ophthalmology. 38 (4): 1365–1378. doi:10.1007/s10792-017-0594-3. ISSN 0165-5701. PMID 28639085. S2CID 4017540.
- ^ Phillips, Robert J et al. "Prostaglandin pathway gene expression in human placenta, amnion and choriodecidua is differentially affected by preterm and term labour and by uterine inflammation." BMC pregnancy and childbirth vol. 14 241. 22 Jul. 2014, doi:10.1186/1471-2393-14-241
- ^ Li Y, He W, Liu T, Zhang Q (December 2010). "A new cyclo-oxygenase-2 gene variant in the Han Chinese population is associated with an increased risk of gastric carcinoma". Mol Diagn Ther. 14 (6): 351–5. doi:10.1007/bf03256392. PMID 21275453. S2CID 1229751.
- ^ Liou JY, Deng WG, Gilroy DW, Shyue SK, Wu KK (September 2001). "Colocalization and interaction of cyclooxygenase-2 with caveolin-1 in human fibroblasts". J. Biol. Chem. 276 (37): 34975–82. doi:10.1074/jbc.M105946200. PMID 11432874.
- ^ Xie WL, Chipman JG, Robertson DL, Erikson RL, Simmons DL (April 1991). "Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing". Proc. Natl. Acad. Sci. U.S.A. 88 (7): 2692–6. Bibcode:1991PNAS...88.2692X. doi:10.1073/pnas.88.7.2692. PMC 51304. PMID 1849272.
Further reading
[edit]- Richards JA, Petrel TA, Brueggemeier RW (February 2002). "Signaling pathways regulating aromatase and cyclooxygenases in normal and malignant breast cells". J. Steroid Biochem. Mol. Biol. 80 (2): 203–12. doi:10.1016/S0960-0760(01)00187-X. PMID 11897504. S2CID 12728545.
- Wu T, Wu H, Wang J, Wang J (2011). "Expression and cellular localization of cyclooxygenases and prostaglandin E synthases in the hemorrhagic brain". J Neuroinflammation. 8: 22. doi:10.1186/1742-2094-8-22. PMC 3062590. PMID 21385433.
- Koki AT, Khan NK, Woerner BM, Seibert K, Harmon JL, Dannenberg AJ, Soslow RA, Masferrer JL (January 2002). "Characterization of cyclooxygenase-2 (COX-2) during tumorigenesis in human epithelial cancers: evidence for potential clinical utility of COX-2 inhibitors in epithelial cancers". Prostaglandins Leukot. Essent. Fatty Acids. 66 (1): 13–8. doi:10.1054/plef.2001.0335. PMID 12051953.
- Saukkonen K, Rintahaka J, Sivula A, Buskens CJ, Van Rees BP, Rio MC, Haglund C, Van Lanschot JJ, Offerhaus GJ, Ristimaki A (October 2003). "Cyclooxygenase-2 and gastric carcinogenesis". APMIS. 111 (10): 915–25. doi:10.1034/j.1600-0463.2003.1111001.x. PMID 14616542. S2CID 23257867.
- Sinicrope FA, Gill S (2004). "Role of cyclooxygenase-2 in colorectal cancer". Cancer Metastasis Rev. 23 (1–2): 63–75. doi:10.1023/A:1025863029529. PMID 15000150. S2CID 21521040.
- Jain S, Khuri FR, Shin DM (2004). "Prevention of head and neck cancer: current status and future prospects". Curr Probl Cancer. 28 (5): 265–86. doi:10.1016/j.currproblcancer.2004.05.003. PMID 15375804.
- Saba N, Jain S, Khuri F (2004). "Chemoprevention in lung cancer". Curr Probl Cancer. 28 (5): 287–306. doi:10.1016/j.currproblcancer.2004.05.005. PMID 15375805.
- Cardillo I, Spugnini EP, Verdina A, Galati R, Citro G, Baldi A (October 2005). "Cox and mesothelioma: an overview". Histol. Histopathol. 20 (4): 1267–74. PMID 16136507.
- Brueggemeier RW, Díaz-Cruz ES (March 2006). "Relationship between aromatase and cyclooxygenases in breast cancer: potential for new therapeutic approaches". Minerva Endocrinol. 31 (1): 13–26. PMID 16498361.
- Fujimura T, Ohta T, Oyama K, Miyashita T, Miwa K (March 2006). "Role of cyclooxygenase-2 in the carcinogenesis of gastrointestinal tract cancers: a review and report of personal experience". World J. Gastroenterol. 12 (9): 1336–45. doi:10.3748/wjg.v12.i9.1336. PMC 4124307. PMID 16552798.
- Bingham S, Beswick PJ, Blum DE, Gray NM, Chessell IP (October 2006). "The role of the cylooxygenase pathway in nociception and pain". Semin. Cell Dev. Biol. 17 (5): 544–54. doi:10.1016/j.semcdb.2006.09.001. PMID 17071117.
- Minghetti L, Pocchiari M (2007). Cyclooxygenase-2, prostaglandin E2, and microglial activation in prion diseases. International Review of Neurobiology. Vol. 82. pp. 265–75. doi:10.1016/S0074-7742(07)82014-9. ISBN 978-0-12-373989-6. PMID 17678966.
External links
[edit]- Nextbio Archived 2019-10-18 at the Wayback Machine
- NSAIDs and Cardiovascular Risk Explained, According to Studies from the Perelman School of Medicine
- Wolfe MM (December 2004). "Rofecoxib, Merck, and the FDA". N. Engl. J. Med. 351 (27): 2875–8, author reply 2875–8. doi:10.1056/NEJM200412303512719. PMID 15625749.