DNA-3-methyladenine glycosylase
| MPG | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | MPG, AAG, ADPG, APNG, CRA36.1, MDG, Mid1, PIG11, PIG16, anpg, N-methylpurine DNA glycosylase | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 156565; MGI: 97073; HomoloGene: 1824; GeneCards: MPG; OMA:MPG - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
DNA-3-methyladenine glycosylase also known as 3-alkyladenine DNA glycosylase (AAG) or N-methylpurine DNA glycosylase (MPG) is an enzyme that in humans is encoded by the MPG gene.[5][6]
Alkyladenine DNA glycosylase is a specific type of DNA glycosylase. This subfamily of monofunctional glycosylases is involved in the recognition of a variety of base lesions, including alkylated and deaminated purines, and initiating their repair via the base excision repair pathway.[7] To date, the human AAG (hAAG) is the only glycosylase identified that excises alkylation-damaged purine bases in human cells.[8]
Function
[edit]DNA bases are subject to a large number of anomalies: spontaneous alkylation or oxidative deamination. It is estimated that 104 mutations appear in a typical human cell per day. Albeit it seems to be an insignificant amount considering the extension of the DNA (1010 nucleotides), these mutations lead to changes in the structure and coding potential of the DNA, affecting processes of replication and transcription.
3-Methyladenine DNA glycosylases are able to initiate the base excision repair (BER) of a wide range of substrate bases that, due to their chemical reactivity, suffer inevitable modifications resulting in different biological outcomes. DNA repair mechanisms take on a vital role in maintaining the genomic integrity of cells from different organisms, in particular 3-Methyladenine DNA glycosylases are found in bacteria, yeast, plants, rodents, and humans. Therefore, there are different subfamilies of this enzyme, such as the Human Alkyladenine DNA Glycosylase (hAAG), that act on other damaged DNA bases apart from 3-MeA.[9]
| tag | AlkA | MAG | mag1 | ADPG | Aag | AGG | aMAG | |
|---|---|---|---|---|---|---|---|---|
| 3-MeA | + | + | + | + | + | + | + | + |
| 3-MeG | + | + | + | + | + | + | ||
| 7-MeG | - | + | + | + | + | + | + | + |
| O2-MeG | - | + | ||||||
| O2-MeC | - | + | ||||||
| 7-CEG | + | + | ||||||
| 7-HEG | + | + | ||||||
| 7-EthoxyG | + | |||||||
| eA | - | + | + | + | + | + | + | |
| eG | + | |||||||
| 8-oxoG | + | + | ||||||
| Hx | - | + | + | + | + | + | + | |
| A | + | + | ||||||
| G | - | + | + | + | + | |||
| T | + | |||||||
| C | + |
Alkylation repairing activity
[edit]In cells,[10] AAG is the enzyme responsible for recognition and initiation of the repair, via catalysing the hydrolysis of the N-glycosidic bond to release the alkylation-damaged purine bases.[11] Specifically, hAAG is able to efficiently identify and excise 3-methyladenine, 7-methyladenine, 7-methylguanine, 1N-ethenoadenine and hypoxanthine.[12]
Deaminated base activity
[edit]MPG can act on all three purine deamination bases: hypoxanthine, xanthine, and oxanine.[13]
Oxanine (Oxa), is a deaminated base lesion in which the N1-nitrogen is replaced by oxygen. Contrary to the alkylation repairing activity, which is only able to act against purine bases, the hAAG is able to excise Oxa[14] from all of four Oxa-containing double stranded base pairs, Cyt/Oxa, Thy/Oxa, Ade/Oxa, and Gua/Oxa, showing no particular preference by any of the bases. In addition hAAG is capable of removing Oxa from single-stranded Oxa- containing DNA. This occurs because the ODG activity of the hAAG does not require a complementary strand.
Structure
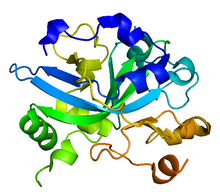
[edit]Alkyladenine DNA glycosylase is a monomeric protein compounded by 298 amino acids, with a formula weight of 33kDa. Its canonical primary structure consists of the following sequence. However, also other functional isoforms have been found.

Isoform 2
[edit]The sequence of this isoform differs from the canonical sequence as follows:
Aminoacids 1-12: MVTPALQMKKPK → MPARSGA
Aminoacids 195-196: QL →HV
Isoform 3
[edit]The sequence of this isoform differs from the canonical sequence in a similar way as the isoform 2:
Aminoacids 1-12: MVTPALQMKKPK → MPARSGA
Isoform 4
[edit]The sequence of this isoform misses the aminoacids 1–17.
It folds into a single domain of mixed α/β structure, with seven α helices and eight β strands. The core of the protein consists of a curved, antiparallel β sheet with a protruding β hairpin (β3β4) that inserts into the minor groove of the bound DNA. A series of α helices and connecting loops form the remainder of the DNA binding interface.[15] It lacks the helix-hairpin-helix motif associated with other base excision-repair proteins and, in fact, it does not resemble any other model in the Protein Data Bank.[15]
Mechanism
[edit]Substrate recognition
[edit]Alkyladenine DNA glycosylase is part of the family of enzymes that follow the BER, acting on specific substrates according to BER steps.
The process of recognition of damaged bases involves initial non-specific binding followed by diffusion along the DNA. Formed the AAG-DNA complex, a redundant process of search occurs because of the long lifetime of this complex, while hAAG search many adjacent sites in a DNA molecule in a single binding. This provides ample opportunity to recognize and excise lesions that minimally perturb the structure of the DNA.[16]
Due to its broad specificity, the hAAG performs the substrate selection through a combination of selectivity filters.[17]
- The first selectivity filter occurs at the nucleotide flipping step of unusable base pairs that present lesions.
- The second selectivity filter is constituted by the catalytic mechanism which ensures that only purine bases are excised, even though smaller pyrimidines can fit in the hAAG's active site. The active site pocket it's designed to accommodate a structurally diverse set of modified purines so it would be difficult to sterically exclude the smaller pyrimidine bases from binding. However, thanks to the different shape and chemical properties of a bound pyrimidine and a purine substrate, the acid-catalyzed reacts only with the pyrimidine preventing it from binding with the hAAG.[10]
- The third selectivity filter consist of unfavorable steric clashes that allow a preferential recognition of purine lesions lacking exocyclic amino groups of guanine and adenine.

Schematic diagram of hAAG-DNA contacts
Nucleotide flipping and fixation
[edit]Its structure contains an antiparallel β sheet with protruding β hairpin (β3β4) that inserts into the minor groove of the bound DNA. This group is unique for the human cells and displaces the selected nucleotide targeted for base excision by flipping it. The nucleotide is secured into the enzyme binding pocket where the active site is found, and is fixed by the amino acids Arg182, Glu125 and Ser262. Also other bonds are formed to bordering nucleotides to stabilize the structure.
The groove in the double helix of DNA left by the flipped-out abasic nucleotide is filled with the lateral chain of the amino acid Tyr162, making no specific contacts with the surrounding bases.

Nucleotide release
[edit]Activated by nearby aminoacids, a water molecule attacks the N-Glycosydic bound releasing the alkylated base via a backside displacement mechanism.
Location
[edit]Human alkyladenine DNA glycosylase localizes to the mitochondria, nucleus and cytoplasm of human cells.[18] Some functionally equivalent enzymes have been found in other species have significantly different structures, such as DNA-3-methyladenine glycosylase in E. coli.[15]
Clinical significance
[edit]According to the mechanism used by Human Alkyladenine DNA Glycosylase, a defect in the DNA repair pathways leads to cancer predisposition. HAAG follows the BER steps so that means that an incorrect role of BER genes could contribute to the development of cancer. Concretely, a bad activity of hAAG may be associated with cancer risk in BRCA1 and BRCA2 mutation carriers.[19]
Aging
[edit]As noted above, DNA-3-methyladenine glycosylase (also called 3-alklyadeneine DNA glycosylase or AAG) is able to identify and excise a variety of alkylation damaged purine bases. Such damages to purine bases occur spontaneously in DNA. Double-mutant mice deficient both for AAG and another enzyme that specifically repairs O6MeG damages (O-6-methylguanine-DNA methyltransferase) had a shorter lifespan and aged more rapidly than wild type mice.[20] These findings indicate that damaged purine bases contribute to the aging process, consistent with the DNA damage theory of aging.
See also
[edit]References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000103152 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000020287 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Chakravarti D, Ibeanu GC, Tano K, Mitra S (Aug 1991). "Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase". The Journal of Biological Chemistry. 266 (24): 15710–5. doi:10.1016/S0021-9258(18)98467-X. PMID 1874728.
- ^ "Entrez Gene: MPG N-methylpurine-DNA glycosylase".
- ^ Hedglin M, O'Brien PJ (2008). "Human alkyladenine DNA glycosylase employs a processive search for DNA damage". Biochemistry. 47 (44): 11434–45. doi:10.1021/bi801046y. PMC 2702167. PMID 18839966.
- ^ Abner CW, Lau AY, Ellenberger T, Bloom LB (Apr 2001). "Base excision and DNA binding activities of human alkyladenine DNA glycosylase are sensitive to the base paired with a lesion". The Journal of Biological Chemistry. 276 (16): 13379–87. doi:10.1074/jbc.M010641200. PMID 11278716.
- ^ Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD (August 1999). "3-methyladenine DNA glycosylases: structure, function, and biological importance". BioEssays. 21 (8): 668–676. doi:10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. PMID 10440863. S2CID 29109365.
- ^ a b O'Brien PJ, Ellenberger T (Oct 2003). "Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines". Biochemistry. 42 (42): 12418–29. doi:10.1021/bi035177v. PMID 14567703.
- ^ Admiraal SJ, O'Brien PJ (Oct 2010). "N-glycosyl bond formation catalyzed by human alkyladenine DNA glycosylase". Biochemistry. 49 (42): 9024–6. doi:10.1021/bi101380d. PMC 2975558. PMID 20873830.
- ^ Hollis T, Lau A, Ellenberger T (Aug 2000). "Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase". Mutation Research. 460 (3–4): 201–10. doi:10.1016/S0921-8777(00)00027-6. PMID 10946229.
- ^ Hitchcock, Thomas M.; Dong, Liang; Connor, Ellen E.; Meira, Lisiane B.; Samson, Leona D.; Wyatt, Michael D.; Cao, Weiguo (2004). "Oxanine DNA Glycosylase Activity from Mammalian Alkyladenine Glycosylase". Journal of Biological Chemistry. 279 (37): 38177–38183. doi:10.1074/jbc.m405882200. ISSN 0021-9258. PMID 15247209.
- ^ Hitchcock TM, Dong L, Connor EE, Meira LB, Samson LD, Wyatt MD, Cao W (Sep 2004). "Oxanine DNA glycosylase activity from Mammalian alkyladenine glycosylase". The Journal of Biological Chemistry. 279 (37): 38177–83. doi:10.1074/jbc.M405882200. PMID 15247209.
- ^ a b c Lau AY, Schärer OD, Samson L, Verdine GL, Ellenberger T (Oct 1998). "Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision". Cell. 95 (2): 249–58. doi:10.1016/S0092-8674(00)81755-9. PMID 9790531. S2CID 14125483.
- ^ Zhang, Yaru (2014). Specificity and Searching Mechanism of Alkyladenine DNA Glycosylase (Thesis). hdl:2027.42/110472.
- ^ Hedglin M, O'Brien PJ (2008). "Human Alkyladenine DNA Glycosylase employs a processive search for dNA damage". Biochemistry. 47 (44): 11434–11445. doi:10.1021/bi801046y. PMC 2702167. PMID 18839966.
- ^ van Loon B, Samson LD (Mar 2013). "Alkyladenine DNA glycosylase (AAG) localizes to mitochondria and interacts with mitochondrial single-stranded binding protein (mtSSB)" (PDF). DNA Repair. 12 (3): 177–87. doi:10.1016/j.dnarep.2012.11.009. hdl:1721.1/99514. PMC 3998512. PMID 23290262.
- ^ Osorio A, Milne RL, Kuchenbaecker K, Vaclová T, Pita G, Alonso R, et al. (Apr 2014). "DNA glycosylases involved in base excision repair may be associated with cancer risk in BRCA1 and BRCA2 mutation carriers". PLOS Genetics. 10 (4): e1004256. doi:10.1371/journal.pgen.1004256. PMC 3974638. PMID 24698998.
- ^ Meira LB, Calvo JA, Shah D, Klapacz J, Moroski-Erkul CA, Bronson RT, Samson LD (September 2014). "Repair of endogenous DNA base lesions modulate lifespan in mice". DNA Repair. 21: 78–86. doi:10.1016/j.dnarep.2014.05.012. PMC 4125484. PMID 24994062.
Further reading
[edit]- Hollis T, Lau A, Ellenberger T (Aug 2000). "Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase". Mutation Research. 460 (3–4): 201–10. doi:10.1016/S0921-8777(00)00027-6. PMID 10946229.
- O'Connor TR, Laval J (May 1991). "Human cDNA expressing a functional DNA glycosylase excising 3-methyladenine and 7-methylguanine". Biochemical and Biophysical Research Communications. 176 (3): 1170–7. doi:10.1016/0006-291X(91)90408-Y. PMID 1645538.
- Samson L, Derfler B, Boosalis M, Call K (Oct 1991). "Cloning and characterization of a 3-methyladenine DNA glycosylase cDNA from human cells whose gene maps to chromosome 16". Proceedings of the National Academy of Sciences of the United States of America. 88 (20): 9127–31. Bibcode:1991PNAS...88.9127S. doi:10.1073/pnas.88.20.9127. PMC 52665. PMID 1924375.
- Kielman MF, Smits R, Devi TS, Fodde R, Bernini LF (1993). "Homology of a 130-kb region enclosing the alpha-globin gene cluster, the alpha-locus controlling region, and two non-globin genes in human and mouse". Mammalian Genome. 4 (6): 314–23. doi:10.1007/BF00357090. PMID 8318735. S2CID 24711956.
- Vickers MA, Vyas P, Harris PC, Simmons DL, Higgs DR (Apr 1993). "Structure of the human 3-methyladenine DNA glycosylase gene and localization close to the 16p telomere". Proceedings of the National Academy of Sciences of the United States of America. 90 (8): 3437–41. Bibcode:1993PNAS...90.3437V. doi:10.1073/pnas.90.8.3437. PMC 46315. PMID 8475094.
- Lau AY, Schärer OD, Samson L, Verdine GL, Ellenberger T (Oct 1998). "Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision". Cell. 95 (2): 249–58. doi:10.1016/S0092-8674(00)81755-9. PMID 9790531. S2CID 14125483.
- Miao F, Bouziane M, Dammann R, Masutani C, Hanaoka F, Pfeifer G, O'Connor TR (Sep 2000). "3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins". The Journal of Biological Chemistry. 275 (37): 28433–8. doi:10.1074/jbc.M001064200. PMID 10854423.
- Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T (Dec 2000). "Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG". Proceedings of the National Academy of Sciences of the United States of America. 97 (25): 13573–8. Bibcode:2000PNAS...9713573L. doi:10.1073/pnas.97.25.13573. PMC 17617. PMID 11106395.
- Daniels RJ, Peden JF, Lloyd C, Horsley SW, Clark K, Tufarelli C, Kearney L, Buckle VJ, Doggett NA, Flint J, Higgs DR (Feb 2001). "Sequence, structure and pathology of the fully annotated terminal 2 Mb of the short arm of human chromosome 16". Human Molecular Genetics. 10 (4): 339–52. doi:10.1093/hmg/10.4.339. PMID 11157797.
- Kim NK, An HJ, Kim HJ, Sohn TJ, Roy R, Oh D, Ahn JY, Hwang TS, Cha KY (2002). "Altered expression of the DNA repair protein, N-methylpurine-DNA glycosylase (MPG) in human gonads". Anticancer Research. 22 (2A): 793–8. PMID 12014652.
- Vallur AC, Feller JA, Abner CW, Tran RK, Bloom LB (Aug 2002). "Effects of hydrogen bonding within a damaged base pair on the activity of wild type and DNA-intercalating mutants of human alkyladenine DNA glycosylase". The Journal of Biological Chemistry. 277 (35): 31673–8. doi:10.1074/jbc.M204475200. PMID 12077143.
- Kim NK, Ahn JY, Song J, Kim JK, Han JH, An HJ, Chung HM, Joo JY, Choi JU, Lee KS, Roy R, Oh D (2003). "Expression of the DNA repair enzyme, N-methylpurine-DNA glycosylase (MPG) in astrocytic tumors". Anticancer Research. 23 (2B): 1417–23. PMID 12820404.
- Watanabe S, Ichimura T, Fujita N, Tsuruzoe S, Ohki I, Shirakawa M, Kawasuji M, Nakao M (Oct 2003). "Methylated DNA-binding domain 1 and methylpurine-DNA glycosylase link transcriptional repression and DNA repair in chromatin". Proceedings of the National Academy of Sciences of the United States of America. 100 (22): 12859–64. Bibcode:2003PNAS..10012859W. doi:10.1073/pnas.2131819100. PMC 240709. PMID 14555760.
- O'Brien PJ, Ellenberger T (Oct 2003). "Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines". Biochemistry. 42 (42): 12418–29. doi:10.1021/bi035177v. PMID 14567703.
- O'Brien PJ, Ellenberger T (Mar 2004). "Dissecting the broad substrate specificity of human 3-methyladenine-DNA glycosylase". The Journal of Biological Chemistry. 279 (11): 9750–7. doi:10.1074/jbc.M312232200. PMID 14688248.
- Likhite VS, Cass EI, Anderson SD, Yates JR, Nardulli AM (Apr 2004). "Interaction of estrogen receptor alpha with 3-methyladenine DNA glycosylase modulates transcription and DNA repair". The Journal of Biological Chemistry. 279 (16): 16875–82. doi:10.1074/jbc.M313155200. PMID 14761960.
- Hitchcock TM, Dong L, Connor EE, Meira LB, Samson LD, Wyatt MD, Cao W (Sep 2004). "Oxanine DNA glycosylase activity from Mammalian alkyladenine glycosylase". The Journal of Biological Chemistry. 279 (37): 38177–83. doi:10.1074/jbc.M405882200. PMID 15247209.
External links
[edit]- DNA-3-methyladenine+glycosylase+II at the U.S. National Library of Medicine Medical Subject Headings (MeSH)