Pairwise Algorithm
A Pairwise Algorithm [1] is an algorithmic technique with its origins in Dynamic programming. Pairwise algorithms have several uses including comparing a protein profile (a residue scoring matrix for one or more aligned sequences) against the three translation frames of a DNA strand, allowing frameshifting. The most remarkable feature of PairWise as compared to other Protein-DNA alignment tools is that PairWise allows frameshifting during alignment.
History
[edit]One of the earliest applications of PairWise to problems in bioinformatics was by Ewan Birney.
Frameshifting refers to the phenomena where in one DNA strands, there are more than one translation frame. For normal Protein-DNA alignment tools, they first choose one of three frames to translate the DNA into a protein sequence, and then compare it with the given protein. Such alignment is based on the assumption that the DNA translation frame is not interrupted for the whole DNA strand. However, this is not generally true.
The PairWise algorithm is a variant of the Smith–Waterman algorithm best local alignment algorithm. These algorithms all belong to the class known as minimal string edit algorithms. The main differences between PairWise and other alignment algorithm is that, besides normal penalties such as Gap Opening Penalty (GOP), Gap Extension Penalty (GEP) and Match, PairWise introduced two new penalties called Frame Opening Penalty (FOP) and Frame Extension Penalty (FEP), which will be incurred when a frameshift is accepted and extended respectively.
Illustration
[edit]Figure 1 illustrates the alignment result when one protein sequence and one DNA sequence was aligned using normal protein-DNA alignment algorithm. The frame used was frame 1 for the DNA sequence. As shown in the picture, there was a gap of 2 amino acids (6 nucleic acids) in the alignment, which results the total low score of -2.

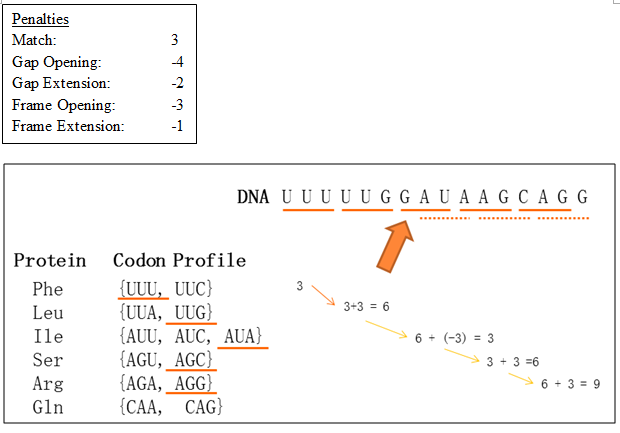
Figure 2 illustrates the aligned result using PairWise. Using the same DNA and protein sequence, and with the penalties modified as below. The arrow is pointing to the position where frameshifting took place. At that nucleotide (G), translation frame was shifted from frame one to frame two (dotted line). This change resulted a much better alignment which has a score of 9.
References
[edit]- ^ Birney, E.; Thompson, J.; Gibson, T. (1996). "PairWise and SearchWise: Finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames". Nucleic Acids Research. 24 (14): 2730–2739. doi:10.1093/nar/24.14.2730. PMC 145991. PMID 8759004.