PrrB/RsmZ RNA family
| PrrB/RsmZ RNA family | |
|---|---|
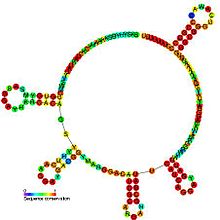 Predicted secondary structure and sequence conservation of PrrB_RsmZ | |
| Identifiers | |
| Symbol | PrrB_RsmZ |
| Rfam | RF00166 |
| Other data | |
| RNA type | Gene; sRNA |
| Domain(s) | Bacteria |
| GO | GO:0006401 GO:0006109 |
| SO | SO:0000655 |
| PDB structures | PDBe |
The PrrB/RsmZ RNA family are a group of related non-coding RNA molecules found in bacteria. PrrB RNA is able to phenotypically complement gacS and gacA mutants and is itself regulated by the GacS-GacA two-component signal transduction system.[1] Inactivation of the prrB gene in Pseudomonas fluorescens F113 resulted in a significant reduction of 2, 4-diacetylphloroglucinol (Phl) and hydrogen cyanide (HCN) production, while increased metabolite production was observed when prrB was overexpressed.[1] The prrB gene sequence contains a number of imperfect repeats of the consensus sequence 5′-AGGA-3′, and sequence analysis predicted a complex secondary structure featuring multiple putative stem-loops with the consensus sequences predominantly positioned at the single-stranded regions at the ends of the stem-loops. This structure is similar to the CsrB and RsmB regulatory RNAs (CsrB/RsmB RNA family),[1] suggesting this RNA also interacts with a CsrA-like protein.
Studies in Legionella pneumophila have shown that the ncRNAs RsmY and RsmZ together with the proteins LetA and CsrA are involved in a regulatory cascade. Also, it appears that these ncRNAs are regulated by RpoS sigma-factor.[2]
See also
[edit]References
[edit]- ^ a b c Aarons S, Abbas A, Adams C, Fenton A, O'Gara F (2000). "A regulatory RNA (PrrB RNA) modulates expression of secondary metabolite genes in Pseudomonas fluorescens F113". J. Bacteriol. 182 (14): 3913–3919. doi:10.1128/JB.182.14.3913-3919.2000. PMC 94573. PMID 10869066.
- ^ Rasis M, Segal G (May 2009). "The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors". Mol. Microbiol. 72 (4): 995–1010. doi:10.1111/j.1365-2958.2009.06705.x. PMID 19400807.
Further reading
[edit]- Duss, O; Michel, E; Diarra dit Konté, N; Schubert, M; Allain, FH (Apr 2014). "Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition". Nucleic Acids Research. 42 (8): 5332–5346. doi:10.1093/nar/gku141. PMC 4005645. PMID 24561806.
