SDHA
| SDHA | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | SDHA, CMD1GG, FP, PGL5, SDH1, SDH2, SDHF, succinate dehydrogenase complex flavoprotein subunit A, MC2DN1, NDAXOA | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 600857; MGI: 1914195; HomoloGene: 3073; GeneCards: SDHA; OMA:SDHA - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Succinate dehydrogenase complex, subunit A, flavoprotein variant is a protein that in humans is encoded by the SDHA gene.[5] This gene encodes a major catalytic subunit of succinate-ubiquinone oxidoreductase, a complex of the mitochondrial respiratory chain. The complex is composed of four nuclear-encoded subunits and is localized in the mitochondrial inner membrane. SDHA contains the FAD binding site where succinate is deprotonated and converted to fumarate. Mutations in this gene have been associated with a form of mitochondrial respiratory chain deficiency known as Leigh Syndrome. A pseudogene has been identified on chromosome 3q29. Alternatively spliced transcript variants encoding different isoforms have been found for this gene.[6]
Structure
[edit]The SDHA gene is located on the p arm of chromosome 5 at locus 15 and is composed of 17 exons.[6] The SDHA protein encoded by this gene is 664 amino acids long and weighs 72.7 kDA.[7][8]
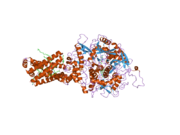
SDHA protein has four subdomains, including capping domain, helical domain, C-terminal domain and most notably, β-barrel FAD-binding domain at N-terminus. Therefore, SDHA is a flavoprotein (Fp) due to the prosthetic group flavin adenine dinucleotide (FAD). Crystal structure suggests that FAD is covalently bound to a histidine residue (His99) and further coordinated by hydrogen bonds with number of other amino acid residues within the FAD-binding domain. FAD which is derived from riboflavin (vitamin B2) is thus essential cofactor for SDHA and whole complex II function.[9]
Function
[edit]
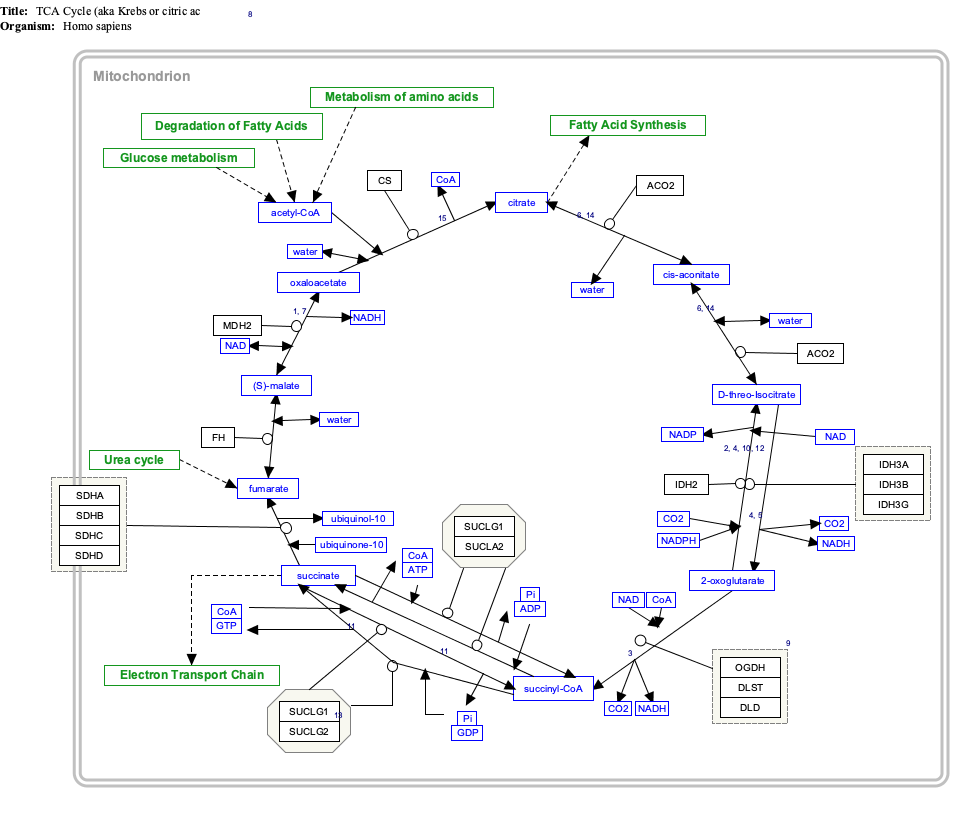
The SDH complex is located on the inner membrane of the mitochondria and participates in both the citric acid cycle and the respiratory chain. The succinate dehydrogenase (SDH) protein complex catalyzes the oxidation of succinate (succinate + ubiquinone => fumarate + ubiquinol). Electrons removed from succinate transfer to SDHA, transfer across SDHB through iron sulphur clusters to the SDHC/SDHD subunits on the hydrophobic end of the complex anchored in the mitochondrial membrane.
Initially, SDHA oxidizes succinate via deprotonation at the FAD binding site, forming FADH2 and leaving fumarate, loosely bound to the active site, free to exit the protein. The electrons derived from succinate tunnel along the [Fe-S] relay in the SDHB subunit until they reach the [3Fe-4S] iron sulfur cluster. The electrons are then transferred to an awaiting ubiquinone molecule at the Q pool active site in the SDHC/SDHD dimer. The O1 carbonyl oxygen of ubiquinone is oriented at the active site by hydrogen bond interactions with Tyr83 of SDHD. The presence of electrons in the [3Fe-4S] iron sulphur cluster induces the movement of ubiquinone into a second orientation. This facilitates a second hydrogen bond interaction between the O4 carbonyl group of ubiquinone and Ser27 of SDHC. Following the first single electron reduction step, a semiquinone radical species is formed. The second electron arrives from the [3Fe-4S] cluster to provide full reduction of the ubiquinone to ubiquinol.[10]
SDHA acts as an intermediate in the basic SDH enzyme action:
- SDHA converts succinate to fumarate as part of the citric acid cycle. This reaction also converts FAD to FADH2.
- Electrons from the FADH2 are transferred to the SDHB subunit iron clusters [2Fe-2S],[4Fe-4S],[3Fe-4S]. This function is part of the Respiratory chain
- Finally the electrons are transferred to the Ubiquinone (Q) pool via the SDHC/SDHD subunits.
Clinical significance
[edit]Because of the complexity of SDHA's locus, SDHA was rarely analyzed,[11] but in an increasing amount of research, it's been found that mutations in SDHA are pathogenic for a number of conditions, including hereditary pheochromocytoma-paraganglioma (PPGL) syndrome, mitochondrial complex II deficiency, gastrointestinal stromal tumors, Leigh syndrome, dilated cardiomyopathy, and possible relation with pituitary adenomas, adrenal carcinomas, and other neuroendocrine tumors.[12] Hereditary PPGL syndrome associated with mutations in SDHA is called "Paragangliomas 5" with likely lower penetrance than other SDHx mutations.[13]
Bi-allelic mutations in SDHA are known to be pathogenic for infant or early childhood Leigh syndrome, a progressive brain disorder.[14][15][16] It is not known, however, how mutations in the SDHA gene are related to the specific features of Leigh syndrome. There is some link between Leigh syndrome as a phenotype of mitochondrial complex II deficiency, but both can occur without the other as relating to SDHA mutations.[17]
SDHA is a tumour suppressor gene, and heterozygous carriers have an increased risk of paragangliomas as well as pheochromocytomas and renal cancer.[18] Risk management for heterozygous carriers of an SDHA mutation typically involve monitoring via annual urine tests for metanephrines and catecholamines as well as non-radiation imaging such as MRIs. PET scans and radiation imaging are used but should be limited to prevent radiation exposure. [19]
Interactive pathway map
[edit]Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- ^ The interactive pathway map can be edited at WikiPathways: "TCACycle_WP78".
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000073578 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000021577 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Hirawake H, Wang H, Kuramochi T, Kojima S, Kita K (July 1994). "Human complex II (succinate-ubiquinone oxidoreductase): cDNA cloning of the flavoprotein (Fp) subunit of liver mitochondria". Journal of Biochemistry. 116 (1): 221–7. doi:10.1093/oxfordjournals.jbchem.a124497. PMID 7798181.
- ^ a b "Entrez Gene: succinate dehydrogenase complex".
- ^ Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, et al. (October 2013). "Integration of cardiac proteome biology and medicine by a specialized knowledgebase". Circulation Research. 113 (9): 1043–53. doi:10.1161/CIRCRESAHA.113.301151. PMC 4076475. PMID 23965338.
- ^ "SDHA - Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial". Cardiac Organellar Protein Atlas Knowledgebase (COPaKB).
- ^ Van Vranken JG, Na U, Winge DR, Rutter J (March–April 2015). "Protein-mediated assembly of succinate dehydrogenase and its cofactors". Critical Reviews in Biochemistry and Molecular Biology. 50 (2): 168–80. doi:10.3109/10409238.2014.990556. PMC 4653115. PMID 25488574.
- ^ Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, et al. (March 2006). "Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction". The Journal of Biological Chemistry. 281 (11): 7309–16. doi:10.1074/jbc.m508173200. PMID 16407191.
- ^ Wagner AJ, Remillard SP, Zhang YX, Doyle LA, George S, Hornick JL (February 2013). "Loss of expression of SDHA predicts SDHA mutations in gastrointestinal stromal tumors". Mod Pathol. 26 (2): 289–94. doi:10.1038/modpathol.2012.153. PMID 22955521.
- ^ "SDHA[gene] ▶ GRCh37". ClinVar. National Library of Medicine.
- ^ van der Tuin K, Mensenkamp AR, Tops CM, Corssmit EP, Dinjens WN, van de Horst-Schrivers AN, Jansen JC, de Jong MM, Kunst HP, Kusters B, Leter EM, Morreau H, van Nesselrooij BM, Oldenburg RA, Spruijt L, Hes FJ, Timmers HJ (February 2018). "Clinical Aspects of SDHA-Related Pheochromocytoma and Paraganglioma: A Nationwide Study". J Clin Endocrinol Metab. 103 (2): 438–445. doi:10.1210/jc.2017-01762. PMID 29177515.
"CORRIGENDUM FOR "Clinical Aspects of SDHA-Related Pheochromocytoma and Paraganglioma: A Nationwide Study"". J Clin Endocrinol Metab. 103 (5): 2077. May 2018. doi:10.1210/jc.2018-00533. PMID 29538659. - ^ "Leigh syndrome". Genetics Home Reference. U.S. National Library of Medicine. Retrieved 30 July 2018.
- ^ Pagnamenta AT, Hargreaves IP, Duncan AJ, Taanman JW, Heales SJ, Land JM, et al. (November 2006). "Phenotypic variability of mitochondrial disease caused by a nuclear mutation in complex II". Molecular Genetics and Metabolism. 89 (3): 214–21. doi:10.1016/j.ymgme.2006.05.003. PMID 16798039.
- ^ Van Coster R, Seneca S, Smet J, Van Hecke R, Gerlo E, Devreese B, et al. (July 2003). "Homozygous Gly555Glu mutation in the nuclear-encoded 70 kDa flavoprotein gene causes instability of the respiratory chain complex II". American Journal of Medical Genetics. Part A. 120A (1): 13–8. doi:10.1002/ajmg.a.10202. PMID 12794685. S2CID 30987591.
- ^ Fullerton M, McFarland R, Taylor RW, Alston CL (2020). "The genetic basis of isolated mitochondrial complex II deficiency". Mol Genet Metab. 131 (1–2): 53–65. doi:10.1016/j.ymgme.2020.09.009. PMC 7758838. PMID 33162331.
- ^ Reference, Genetics Home. "SDHA". Genetics Home Reference. Retrieved 2016-08-31.
- ^ "SDHA, SDHB or SDHC-related familial paraganglioma-phaeochromocytoma – risk management". eviQ Cancer Treatments Online. Cancer Institute NSW. 22 February 2022. 4066 v.3.
Further reading
[edit]- Aboulaich N, Vainonen JP, Strålfors P, Vener AV (October 2004). "Vectorial proteomics reveal targeting, phosphorylation and specific fragmentation of polymerase I and transcript release factor (PTRF) at the surface of caveolae in human adipocytes". The Biochemical Journal. 383 (Pt 2): 237–48. doi:10.1042/BJ20040647. PMC 1134064. PMID 15242332.
- Bonache S, Martínez J, Fernández M, Bassas L, Larriba S (June 2007). "Single nucleotide polymorphisms in succinate dehydrogenase subunits and citrate synthase genes: association results for impaired spermatogenesis". International Journal of Andrology. 30 (3): 144–52. doi:10.1111/j.1365-2605.2006.00730.x. PMID 17298551.
- Horváth R, Abicht A, Holinski-Feder E, Laner A, Gempel K, Prokisch H, et al. (January 2006). "Leigh syndrome caused by mutations in the flavoprotein (Fp) subunit of succinate dehydrogenase (SDHA)". Journal of Neurology, Neurosurgery, and Psychiatry. 77 (1): 74–6. doi:10.1136/jnnp.2005.067041. PMC 2117401. PMID 16361598.
- Kullberg M, Nilsson MA, Arnason U, Harley EH, Janke A (August 2006). "Housekeeping genes for phylogenetic analysis of eutherian relationships". Molecular Biology and Evolution. 23 (8): 1493–503. doi:10.1093/molbev/msl027. PMID 16751257.
- Tomitsuka E, Kita K, Esumi H (2009). "Regulation of succinate-ubiquinone reductase and fumarate reductase activities in human complex II by phosphorylation of its flavoprotein subunit". Proceedings of the Japan Academy. Series B, Physical and Biological Sciences. 85 (7): 258–65. Bibcode:2009PJAB...85..258T. doi:10.2183/pjab.85.258. PMC 3561849. PMID 19644226.
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, et al. (August 2009). "SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma". Science. 325 (5944): 1139–42. Bibcode:2009Sci...325.1139H. doi:10.1126/science.1175689. PMC 3881419. PMID 19628817.
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, et al. (January 2006). "Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes". Genome Research. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- González-Cabo P, Vázquez-Manrique RP, García-Gimeno MA, Sanz P, Palau F (August 2005). "Frataxin interacts functionally with mitochondrial electron transport chain proteins". Human Molecular Genetics. 14 (15): 2091–8. doi:10.1093/hmg/ddi214. PMID 15961414.
- Baysal BE, Lawrence EC, Ferrell RE (March 2007). "Sequence variation in human succinate dehydrogenase genes: evidence for long-term balancing selection on SDHA". BMC Biology. 5: 12. doi:10.1186/1741-7007-5-12. PMC 1852088. PMID 17376234.
- Eng C, Kiuru M, Fernandez MJ, Aaltonen LA (March 2003). "A role for mitochondrial enzymes in inherited neoplasia and beyond". Nature Reviews. Cancer. 3 (3): 193–202. doi:10.1038/nrc1013. PMID 12612654. S2CID 20549458.
- Brière JJ, Favier J, El Ghouzzi V, Djouadi F, Bénit P, Gimenez AP, Rustin P (October 2005). "Succinate dehydrogenase deficiency in human". Cellular and Molecular Life Sciences. 62 (19–20): 2317–24. doi:10.1007/s00018-005-5237-6. PMC 11139140. PMID 16143825. S2CID 23793565.
- Korsten A, de Coo IF, Spruijt L, de Wit LE, Smeets HJ, Sluiter W (February 2010). "Patients with Leber hereditary optic neuropathy fail to compensate impaired oxidative phosphorylation". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1797 (2): 197–203. doi:10.1016/j.bbabio.2009.10.003. PMID 19836344.
- Hendrickson SL, Lautenberger JA, Chinn LW, Malasky M, Sezgin E, Kingsley LA, et al. (September 2010). "Genetic variants in nuclear-encoded mitochondrial genes influence AIDS progression". PLOS ONE. 5 (9): e12862. Bibcode:2010PLoSO...512862H. doi:10.1371/journal.pone.0012862. PMC 2943476. PMID 20877624.
- Van Coster R, Seneca S, Smet J, Van Hecke R, Gerlo E, Devreese B, et al. (July 2003). "Homozygous Gly555Glu mutation in the nuclear-encoded 70 kDa flavoprotein gene causes instability of the respiratory chain complex II". American Journal of Medical Genetics. Part A. 120A (1): 13–8. doi:10.1002/ajmg.a.10202. PMID 12794685. S2CID 30987591.
- Wang L, McDonnell SK, Hebbring SJ, Cunningham JM, St Sauver J, Cerhan JR, et al. (December 2008). "Polymorphisms in mitochondrial genes and prostate cancer risk". Cancer Epidemiology, Biomarkers & Prevention. 17 (12): 3558–66. doi:10.1158/1055-9965.EPI-08-0434. PMC 2750891. PMID 19064571.
- Huang G, Chen Y, Lu H, Cao X (February 2007). "Coupling mitochondrial respiratory chain to cell death: an essential role of mitochondrial complex I in the interferon-beta and retinoic acid-induced cancer cell death". Cell Death and Differentiation. 14 (2): 327–37. doi:10.1038/sj.cdd.4402004. PMID 16826196.
- Sifroni KG, Damiani CR, Stoffel C, Cardoso MR, Ferreira GK, Jeremias IC, et al. (September 2010). "Mitochondrial respiratory chain in the colonic mucosal of patients with ulcerative colitis". Molecular and Cellular Biochemistry. 342 (1–2): 111–5. doi:10.1007/s11010-010-0474-x. PMID 20440543. S2CID 34103232.
- Ma YY, Wu TF, Liu YP, Wang Q, Li XY, Ding Y, et al. (May 2014). "Two compound frame-shift mutations in succinate dehydrogenase gene of a Chinese boy with encephalopathy". Brain & Development. 36 (5): 394–8. doi:10.1016/j.braindev.2013.06.003. PMID 23849264. S2CID 24730430.