Low Frequency of Microsatellites in the Avian Genome
Abstract
A better insight into the occurrence of microsatellites in a range of taxa may help to understand the evolution of simple repeats. Previous studies have found the relative abundance of several repeat motifs to differ among mammals, invertebrates, and plants. Absolute numbers of microsatellites also tend to correlate positively with genome size. We analyzed the occurrence, frequency, and distribution of microsatellites in birds, a taxon with one of the smallest known genome sizes among vertebrates. Dot-blot hybridization revealed that about half of 22 different di-, tri-, and tetranucleotide repeat motifs were clearly more common in human than in three species of birds: chicken, woodpecker, and swallow. For the remaining motifs no clear difference was found. From searching avian database sequences we estimated there to be 30,000–70,000 microsatellites longer than 20 bp in the avian genome. The number of (CA)⩾10 would be around 7000–9000 and the number of (CA)⩾14 about 3000. The calculated density of avian microsatellites (total, one every 20–39 kb; (CA)⩾10, one every 136–150 kb) is much lower than that estimated for the human genome (one every 6 and 30 kb, respectively). This may be explained by the fact that the avian genome contains relatively less noncoding DNA than most mammals and that avian SINE/LINE elements do not terminate in poly(A) tails, which are known to provide a resource for the evolution of simple repeats in mammals. We found no association between microsatellites and SINEs in birds. Primed in situ labeling suggested fairly even distribution of (CA)n repeats over chicken macrochromosomes and intermediate chromosomes, whereas the microchromosomes, a large part of the Z and W chromosomes, and most telomeres and centromeres had very low concentrations of (CA)n microsatellites. The scarcity of microsatellites on the microchromosomes is compatible to these regions likely being unusually rich in coding sequences. The low microsatellite density in the genome in general and on the microchromosomes in particular imposes an obstacle for the development of marker-rich genetic maps of chicken and other birds, and for the localization of quantitative trait genes.
Less than 10 years after their introduction, microsatellites have developed into the marker of choice in a number of genetic areas, including genome mapping and medical, evolutionary, and ecological genetics. Despite this widespread use, many questions still remain unresolved regarding the evolution of these simple, repeated sequences, not least the mutational processes involved (Amos et al. 1996; Primmer et al. 1996). Such understanding is important for the proper use of microsatellites in evolutionary contexts, for example, for phylogeny reconstruction (Jarne and Lagoda 1996) and also for addressing general questions relating to genome organization. Knowledge of the patterns of microsatellite evolution may also help to reveal whether these sequences are associated with a functional significance. One intriguing question in this respect is why certain repeat motifs are more common than others, and why this varies among taxa. In human, (A)n and (CA)n are by far the most common motif variants, the latter being the most widely studied marker. Although a similar situation exists in other mammals (e.g.,Beckmann and Weber 1992), the same pattern is not true for all taxa. In at least some insect species, (CT)n is more common than (CA)n (Estoup et al. 1993) as is the case for an oyster species Ostrea edulis (Naciri et al. 1995). In plants, (AT)n, which is rare in mammals, clearly outnumbers (CA)n (Lagercrantz et al. 1993).
Related to evolution and a possible functional role, comprehensive studies of the absolute numbers of microsatellites in various genomes are needed to comparatively address whether microsatellite abundance is a direct function of genome size. Microsatellites predominantly occur in noncoding DNA, and if the amount of such sequences acts as the sole constraint for the evolution of simple repeats, the absolute numbers of repeats in the genome should correlate closely to DNA content. That this is at least probable has been indicated from hybridization experiments with divergent taxa (Hamada et al. 1982). Furthermore, bats, which have one of the smallest genomes among mammals, appear to harbor considerably fewer microsatellites than other mammalian species (Van Den Bussche et al. 1995).
In this study we have analyzed the occurrence, frequency, and distribution of microsatellites in birds. Avian genomes are generally small; the domestic chicken (Gallus gallus) genome, for instance, is estimated to contain close to one-third the number of base pairs of that in the human genome (Bloom et al. 1993; Wachtel and Tiersch 1993). It would thus be expected that avian genomes harbor significantly fewer microsatellites than most mammals, and there are indications that this may be the case for dinucleotide motifs (Hamada et al. 1982; Manor et al. 1988). Our data from dot-blot hybridizations and database searches now show conclusively that microsatellite repeats generally occur less frequently in birds compared to other vertebrates, and in contrast to mammalian microsatellites, bird microsatellites do not appear to be associated with short interspersed repetitive elements (SINEs).
RESULTS
Dot-Blot Hybridization
To compare the abundance of different repeat motifs in avian and mammalian genomes we performed dot-blot hybridizations with equimolar amounts of chicken, white-backed woodpecker Dendrocopus leucotos, barn swallow Hirundo rustica, pig, and human DNA. The three bird species were chosen so as to represent three divergent avian lineages; human and pig were included as reference mammalian species. A large set of hybridization probes was applied, including 2 dinucleotide repeats [(CA)15 and (CT)15], all 10 possible trinucleotide repeats [(AAT)10, (AAC)10, (AAG)10, (AGT)10, (GAT)10, (ACG)10, (CAG)10, (CCT)10, (GGT)10, (CCG)10], and 9 tetranucleotide repeats [(AAAT)15, (AAAC)7, (AAAG)7, (CATT)7, (GAAT)7, (GATA)7, (GACA)7, (GGAA)7, and (GGAT)7].
In general, hybridization signals from mammalian DNA were stronger than signals from avian samples, whereas there were only slight variations between the signal intensities obtained from the two mammalian and the three avian species (Fig. 1). We scanned the hybridization filters using a PhosphorImager to quantify these differences and derived hybridization intensity ratios for human/bird signals (Table 1). Dinucleotide repeats were particularly less frequent in birds, with the CA and CT motifs both being 10–15 times more common in human. Some tri- and tetranucleotide repeat motifs (AAC, ACC, AAAT, AAGT, and AGAT) were similarly biased toward a considerably higher frequency in human. For most of the remaining types of repeats the human/bird intensity ratio fell between 0.5 and 2, which we do not consider to be clear evidence for significantly different frequencies in the two taxa. Only 3 of 22 human/pig ratios fell outside the 0.5–2.0 range, with two motifs being more common in human (AAG ratio = 3.6; AAAT ratio = 2.3) while AAC was more than twice as common in the porcine genome (ratio = 0.4).
Dot-blot hybridization signals of (CA)15 (a), (AAC)10 (b), and (AAGG)7 (c) to genomic DNA of human (1), pig (2), chicken (3), woodpecker (4), and swallow (5). Intensity ratio calculations from these filters suggested (CA)15 and (AAC)10 to be more common in the mammalian than in the avian genomes, whereas (AAGG)7 appeared to occur equally in all species (refer to Table 1). The uppermost dot on each filter contains 300, 280, 120, 120, and 120 ng of genomic DNA, respectively. The remaining three dots represent 50%, 25%, and 12.5% dilutions of the above concentrations. Different exposure times were used for the filters; therefore signal intensities between filters cannot be compared.
Intensity Ratios (Human:Bird) of Hybridization Signals to Genomic DNA Probed with Various Microsatellite Motifs
For the most commonly used microsatellite marker (CA)n we also employed dot-blot hybridization to estimate the absolute number of loci present in the chicken genome. Because this estimate is highly dependent on the minimum length of repeats that are detected by the probe, we first determined the lower limit of repeat length uncovered by the hybridization stringency applied. This was accomplished by including a panel of plasmid DNAs with cloned (CA)n repeats of different length: (CA)10, (CA)12, (CA)14, (CA)16, and (CA)18. Although little or no signal was obtained with (CA)10 and (CA)12 plasmid DNAs, hybridization intensities for (CA)14, (CA)16, and (CA)18 were about the same (data not shown). From this we concluded that hybridization stringency was adjusted to the detection of repeat lengths of (CA)14 and longer. By relating hybridization intensities of genomic and plasmid DNAs we subsequently estimated there to be ∼1500 copies of (CA)⩾14 in the chicken genome. The corresponding estimates for human and pig DNA were 17,000 and 16,000, respectively.
Survey of Database Sequences
There was ∼3.5 Mb of avian nonmitochondrial sequence in Genbank release 93, with the majority constituting chicken sequences. We searched these 2933 sequences for the presence of microsatellite repeats with the criterion of a perfect repetitive array of at least 20 bp in length, that is, the same search routine as used by Beckmann and Weber (1992) analyzing human repeats. In total, 117 unique bird microsatellites were identified, yielding an average density of 1 every 31 kb (Table 2). This density is clearly lower than one every 6 kb reported for human DNA, but the avian estimate may be flawed, as discussed below. The most common avian repeat type was (A)n (26 hits) followed, somewhat surprisingly by (AGG)n (15 hits) and then (CA)n(10), (CCG)n (9), (AAAC)n, and (AAAT)n (8). Most repeats however, were rather short, and (A)n and (CA)n clearly outnumbered all other repeat motifs if only considering loci with at least 10 repeat units (Table 2).
Number of Avian Microsatellites in the GenBank 93 Database
A closer dissection of database sequences indicated that the observed frequencies of different microsatellites may not be representative for the avian (chicken) genome. Avian Genbank entries are significantly biased toward coding sequences, with almost two-thirds of sequences being cDNA clones. When one also considers the proportion of DNA in remaining genomic clones that constitute coding sequences, the total fraction of coding DNA among avian Genbank entries is >60%, a considerably higher proportion than in the genome as a whole. The fact that most microsatellites should be expected to be situated in noncoding DNA suggests therefore that the database screening underestimated the number of bird microsatellites. In support of this, different repeat types showed marked differences with regard to their location within coding or noncoding DNA (Table 3). Importantly, mono-, di-, and tetranucleotide repeats were totally excluded from coding DNA, whereas trinucleotide repeats were found regularly within coding sequences. Therefore, trinucleotide repeats may be over-represented in our sample of database hits.
Distribution of Repeat Classes in Different DNA Regions
We used two approaches to derive appropriate estimates of the absolute numbers of microsatellites in the avian genome. First, similar to Beckman and Weber (1992), we extracted long (>10-kb) genomic clones and tallied the number of microsatellites. Seven repeats found within the 272 kb of DNA contained in 15 such clones give an estimated frequency of one repeat every 39 kb, or a total of ∼31,000 loci in the genome. This density, though obviously very approximate, is significantly lower than that calculated by Beckmann and Weber (1992)for the human genome (χ2 = 16.1,P = 0.0001; df = 1; 122 microsatellites in 745 kb). Second, we sought to convert the frequencies of avian microsatellites in Genbank to that expected for the whole genome by compensating for the overrepresentation of expressed sequences in the database. This can be done using the simple expressionD = 1/[af c + (1 − a)f nc], where D is the genomic density (one microsatellite everyD bp), a is the proportion of coding DNA in the genome and f c and f nc are the microsatellite frequencies (per bp) in coding and noncoding DNA, respectively, as determined from database occurrence (see Methods). A problem in this context is to derive an estimate of what proportion of the avian genome is made up of coding DNA. In mammals, a figure of 10% is often assumed (Hochgeschwender and Brennan 1991), but as birds have only about one-third of the mammalian DNA content, avian genes may constitute as much as 30% of the genome. Using gene proportions of both 10% and 30%, the total number of microsatellites ⩾20 bp was estimated at 60,000–73,000 (one every 16–20 kb; Table4). The number of (CA)⩾10 copies would be 6600–8500, or one every 140–180 kb. Although data are sparse and must be treated cautiously, the number of (CA)⩾14 would be 2600–3400 (one every 350–450 kb). Interestingly, both database approaches used for estimating total microsatellite numbers in the avian genome gave similar figures, suggesting a markedly lower density than in the human genome. Furthermore, the estimated number of avian (CA)⩾14 loci was similar to that obtained from the dot-blot hybridization experiments.
Estimated Numbers of Microsatellites ⩾20 bp in Length in the Avian Genome Assuming a Gene Content of 10% or 30% per Genome
No Association with Other Repetitive Elements
Various microsatellites in other taxa are known to be directly associated with SINEs (e.g., Beckmann and Weber 1992) and may even have evolved from sequences contained in such elements (Arcot et al. 1995;Nadir et al. 1996). We therefore investigated whether this type of association is also found in birds. The predominant SINE in chicken is called CR1 (Stumph et al. 1981), and estimates of its copy number range from 7,000 to 20,000 (Hache and Deeley 1988) to >100,000 (Vandergon and Reitman 1994). In 81 genomic sequences harboring CR1 elements (Vandergon and Reitman 1994) we found 14 that also harbored microsatellites. In none of these was the microsatellite directly associated with the CR1 element. In fact only one (A)n repeat was located 25 bp 3′ of a CR1 element whereas all other 13 microsatellite repeats were situated farther than 350 bp, 5′ or 3′, from the SINE. Moreover, a database search with, on average, 40 bp of DNA flanking each side of all 117 identified microsatellites failed to find significant homology with known repetitive elements, avian or otherwise, or with each other, which also indicates that any association between microsatellites and SINEs in birds would, at best, be rare.
Primed In Situ Labeling
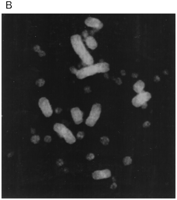
We also investigated whether the apparent lower density of avian microsatellites could be explained by an unusual chromosomal distribution. Primed in situ (PRINS) labeling (Koch et al. 1989) of chicken metaphase chromosome spreads with a (CA)10 primer suggested, however, a relatively even distribution of (CA)n repeats over the macrochromosomes and the intermediate chromosomes, with slight variation in the intensity of fluorescent signals, as exemplified in Figure2. To get a proper picture of how signal intensities varied over chromosomes, hybridization signals from 40 metaphase spreads were quantified by dividing the larger autosomes and the sex chromosomes into equal sized discrete regions. This was not possible for the intermediate chromosomes and microchromosomes, as they are extremely difficult to identify by cytological means. Neither were we able to distinguish the two chromosomal arms of the nearly metacentric W chromosome. The signal in each region was classified as weak, medium, or strong (Fig. 3). Several interesting patterns emerged. First, pooled data from all microchromosomes revealed that they generally hybridized less intensely. Second, the telomeric regions of the macrochromosomes also hybridized less brightly than other chromosomal regions. This was also true for most centromeres, although this is not manifested in Figure 3 because of the classified intervals containing centromeres generally being too large to allow such a resolution. Third, although mostly showing only weak signals over large parts of the chromosomes, the telomeric part of the p arm of the Z chromosome and, similarly, the end of one of the chromosomal arms of the W chromosome showed intense signals. Therefore, these data indicate a relatively low frequency of (CA)n microsatellites on the microchromosomes, on most of the Z and W chromosomes and on telomeres and centromeres.
PRINS hybridization of a (CA)10 oligonucleotide to a female chicken metaphase spread. (A) Chromosomes were counterstained with DAPI. (B) (CA)10 hybridization in the absence of DAPI staining
Quantification of PRINS (CA)10 hybridization to 40 chicken metaphase spreads. Signal intensities for the five largest chromosomes, the sex chromosome, and the microchromosomes as a whole were classified using a point system as either weak (0), average (+1), or strong (+2). These values were transformed into percentages for defined regions of each chromosome so that, e.g., a classification of average for a particular chromosomal region on all metaphases correlates to a value of 50%. Signals from the W chromosome are shown with the assumption that the bright signal on the end of one of the chromosomal arms was consistently from the end of the p arm (see text).
DISCUSSION
Density of Avian Microsatellites
Dot-blot hybridization, as well as the incidence of repeat sequences in database entries, suggests microsatellites to be generally less frequent in birds than in mammals. For instance, 11 of 22 di-, tri-, and tetranucleotide repeat motifs gave considerably weaker dot-blot hybridization signals to avian than to human or porcine DNA, whereas no probe motif showed the opposite trend. Based on database screening, we estimated the total number of microsatellites ⩾20 bp in the avian genome to be 60,000–73,000, or 1 every 16–20 kb and if only genomic clones ⩾10 kb were considered, the estimated number of avian repeats was only 31,000 (1 every 39 kb). The number of (CA)⩾10 copies would be 6600–8500 (1 every 140–180 kb), and the number of (CA)⩾14 loci would be 2600–3400 (1 every 350–450 kb). Dot-blot hybridization gave an estimate of 1500 loci for the latter repeat type. The higher densities are reached if the proportion of genes in the avian genome is 10%, as assumed for many mammals, whereas the lower densities can be estimated if genes constitute 30% of the avian genome. A figure of 30% seems more plausible, as the mean avian genome size is only 1.2 × 109 bp (Tiersch and Wachtel 1991; Bloom et al. 1993). This also depends on whether birds have about the same number of genes as mammals and if mean gene size is similar in the two taxa. Comparisons of exon lengths in orthologous genes (Hughes and Hughes 1995) lend support to a similar gene size in human and birds. It should be noted that our estimated microsatellite frequencies were obtained mainly from chicken sequence data. The similarity of avian hybridization intensities for most repeat motifs (Table 1) and the remarkable consistency of bird species’ DNA content (Tiersch and Wachtel 1991) indicate that these estimated repeat frequencies may also be representative for other birds.
The smaller genome size of birds compared to mammals would logically predict avian genomes to contain lower absolute numbers of microsatellites than, for example, human, as was the case. However, the number of microsatellites in the human compared to the avian genome still exceeds this expectation greatly. Although our dot-blot data suggested that there are 1500 (CA)⩾14 copies in chicken, the number in human was at least 10 times more, ∼17,000. Our human estimate is considerably less than the 50,000–100,000 estimated in earlier studies (e.g., Hamada et al. 1982; Tautz and Renz 1984;Beckmann and Weber 1992), but these kind of estimates are highly dependent on the length of repeats detected during hybridization and it is evident that at least some of the previous studies of human DNA have included (CA)n loci with <14 repeat units. Our database screenings gave further support for the nonproportional excess of human microsatellites. We estimated the total microsatellite density to be one every 39 kb in birds, whereas Beckmann and Weber (1992)obtained an estimate of one every 6 kb in humans using similar search procedures. Similarly, our estimate of one (CA)⩾10 copy every 136 kb in birds is clearly lower than one every 30 kb estimated in human (Beckmann and Weber 1992). The general low frequency of avian microsatellites found in this study is compatible with circumstantial observations of low numbers of avian dinucleotide repeats found previously (Hamada et al. 1982; Manor et al. 1988; Moran 1993; Cheng et al. 1995).
Is the fact that birds seem to be devoid of microsatellites adaptive, or is this just a matter of coincidence? This question should take into account the low levels of interspersed repetitive DNA observed in birds (∼30%; Epplen et al. 1978; Eden and Hendrick 1978; Venturini et al. 1987) and the shorter average size of avian introns compared with human homologs (Hughes and Hughes 1995). Taken together, this suggests that there is (or has been) a constraint on avian genome size operating. It has been speculated that the physical requirements related to flight may place limits on genome size and therefore on the amount of nonfunctional DNA tolerable (Cavalier-Smith 1978; Hughes and Hughes 1995). In support of this theory, bats have one of the smallest known genome sizes of all mammals (Burton et al. 1989) and have also a reduced frequency of microsatellites (Van Den Bussche et al. 1995).
If one assumes that there is a constraint on avian genome size, it still remains to be explained why the density of microsatellites is low in birds. A higher gene density in birds compared with mammals would imply that the relative amount of noncoding DNA should be less in birds, in turn giving less opportunities for avian microsatellites to evolve. This could be accentuated if unique noncoding DNA is favored over repetitive DNA in birds, a possible scenario if noncoding DNA acts at least partly as raw material for the evolution of novel genes. If so, it remains to be clarified if the low tempo of microsatellite evolution is governed by factors involved in the generation of simple repeats by replication slippage or solely by direct selection against “superfluous” repetitive DNA.
Another situation that may contribute to low frequencies of microsatellites in birds is the apparent lack of association between avian SINEs/LINEs (long interspersed repetitive elements) and microsatellites as compared to mammals, where the two types of repetitive elements are commonly found adjacent to each other (Economou et al. 1990; Beckmann and Weber 1992; Buchanan et al. 1993; Ellegren 1993; Varvio and Kaukinen 1993). Mammalian SINEs (and LINEs) regularly terminate in a poly(A) tail derived from retrotransposition, and these stretches provide the basis for the evolution of more complex A-rich repeats like (A1–3N)n (Arcot et al. 1995;Duffy et al. 1996; Nadir et al. 1996). Because SINEs and LINEs appear to be less abundant in birds than in mammals (Deininger 1989; Hutchison et al. 1989) and, importantly, because those avian interspersed elements characterized so far do not terminate with a poly(A) tail (e.g., Chen et al. 1995), low frequencies of microsatellites in birds may be at least partly attributable to a lack of poly(A) tails of interspersed elements providing a source for transitions into various types of repeats.
Distribution of (CA)n Loci on Chicken Chromosomes
PRINS labeling of chicken metaphase chromosomes using a (CA)10 primer suggested a relatively even distribution of (CA)n loci across the five largest chromosomes and the intermediate chromosomes. The only exceptions were the centromeres and telomeres, which consistently stained less brightly on all chromosomes. Centromeres and telomeres are known from a wide variety of species to be built up by other specific repeats (e.g., Biessmann and Mason 1992), and the scarcity of (CA)nmicrosatellites in these regions is therefore not surprising. A lack of (CA)n repeats has also been observed on porcine centromeres and telomeres using the same technique (Winterø et al. 1992). Most of the female-specific W chromosome (birds have a reversed sex chromosome system as compared with mammals, with females being heterogametic ZW and males being homogametic ZZ) also gave rather faint signals with only one of the chromosomal ends staining brightly. We were not able to distinguish the two arms of the W chromosome, but the fact that we never observed a W chromosome with intense signals at both ends suggests that it consistently was the same end that labeled brightly. Some 75% of the chicken W chromosome constitutes huge arrays of satellite DNAs of a few repeat motifs (Saitoh and Mizuno 1992). These fill up the entire q arm and the proximal half of the p arm. Accordingly, the W chromosome harbors very few coding sequences, and the first avian W-linked gene has been identified only recently (Ellegren 1996; Griffiths et al. 1996). The fact that only the distal part of the p arm appears to contain unique DNA (Saitoh and Mizuno 1992) agrees very well with our detection of (CA)nloci on only one end of the chromosome, which hence should be expected to be the telomeric part of the p arm. The few genes present on the W chromosome should similarly be expected to reside in this region. The observation that the (CA)10 probe hybridized more intensely to the telomeric part of the p arm of the Z chromosome than to the remaining part of the chromosome is more surprising. Perhaps the similar distribution of (CA)n loci on the Z and W chromosomes reflects a common ancestry.
Generally faint signals from the microchromosomes suggested (CA)n density to be low on these tiny DNA segments. An alternative explanation could possibly be that the small size of the chromosomes prevented strong signals from emerging to some extent; however, we do not think this was the case because careful examination of 40 metaphases did not reveal any strong signal from any microchromosome. It has become evident from the assignments of a number of genes that the microchromosomes do not represent genetically inert reserves of DNA (e.g., Bloom and Bacon 1985), a situation assumed previously. McQueen et al. (1996) have shown recently that there are a disproportionately high number of CpG islands, indicative of a high gene content, on the chicken microchromosomes. The average density may be as high as one gene per 10 kb, an exceptional value as compared with genome organization in mammals. This high concentration of genes coupled with a generally shorter avian intron size (Hughes and Hughes 1995) would imply reduced possibilities for the evolution of simple repeats from noncoding DNA on the microchromosomes. It should be noted, however, that the lower microsatellite density in the avian genome cannot be attributed entirely to their sparsity on the microchromosomes as they only account for 25% of the DNA in the avian genome (Bloom et al. 1993).
Practical Implications for Avian Genome Analysis
Regardless of the reason behind the lower frequency of microsatellites in birds, this factor will limit the ability to develop marker-rich genetic maps to the extent of those achieved in mammalian species. The lack of association with SINEs/LINEs helps to but does not entirely cancel out this negative effect, as it should be possible to derive locus-specific amplification of a higher percentage of microsatellites than it is when these microsatellites are immediately flanked by other repetitive elements (Economou et al. 1990). As there does not appear to be a single microsatellite motif in birds as common as (CA)n in mammals, it would be advisable to search with a range of repeat motifs in library screenings. The low density of avian microsatellites also warrants the use of marker-enriched libraries (e.g., Armour et al. 1994) to avoid screening extensive numbers of clones from bulk libraries. Another practical consequence is that cloning the same marker more than once will become increasingly common (Cheng et al. 1995), highlighting the importance of screening sequence databases before developing PCR primers for new clones.
The particularly low microsatellite density on microchromosomes, together with a contrasting high gene density, introduces a special obstacle for genetic mapping in chicken and other birds. Chicken microchromosomes possess an obligate chiasmata, meaning that the genetic length of each chromosome is 50 cM. The 29 chicken microchromosomes contain ∼300 Mb (Bloom et al. 1993), and the average ratio of genetic to physical distance would thus be 5 cM per megabase of DNA. This is an extremely skewed figure, paralleling that recorded in telomeric regions of mammalian chromosomes (e.g.,Hultén 1974). Frustratingly, the chromosomal regions likely associated with the highest gene contents also coincide with regions of unusually high recombination frequencies but with reduced numbers of potential genetic markers. Coupled with the difficulty in using comparative gene map data from phylogenetically divergent mammals like human and mouse, the localization of trait genes by positional (candidate) cloning will not be easy for avian geneticists.
METHODS
Dot-Blot Hybridization
Genomic DNA was prepared by proteinase K digestion, phenol extraction, and ethanol precipitation from blood of three avian species belonging to different superorders: the domestic chicken, the white-backed woodpecker, and the swallow and from human and large white domestic pig. The quality of the different DNA preparations was similar, as judged from A 260/A 280ratios. The amount of genomic DNA used on dot-blot filters was calculated taking into account the differing genome sizes so that the quantity of DNA used for each species contained approximately the same number of genome copies (equimolar amounts). Starting from 300 ng of human DNA (assuming a genome size of 3.0 × 109 bp), 280 ng of pig DNA (2.8 × 109 bp; Schmitz et al. 1992) and 120 ng of DNA of the three avian species (1.2 × 109 bp;Bloom et al. 1993), serial dilutions of 50%, 25%, and 12.5% were also analyzed to minimize potential experimental biases (cf.Anchordoguy et al. 1996).
Twelve identical Hybond-N+ filters (Amersham, UK) containing DNA as indicated above were prepared following the manufacturer’s instructions by manually dotting DNA solutions using recently calibrated pipettes. Filters were prehybridized for 1 hr in a solution of 7% SDS, 1 mm EDTA, 1% BSA, 0.26 mNa2HPO4, and 10% dextran sulfate at 65°C, after which 40 pmoles of single-stranded oligonucleotide, previously end-labeled using 1.6 μCi of [γ-32P]ATP, was added to the solution and hybridization was allowed to continue overnight. Filters were then washed three times in 0.3× SSC solution; once for 5 min at room temperature, followed by two 15-min washes at the following temperatures (calculated melting temperature of oligonucleotide in 0.3× SSC 5°C): (AAT)10 at 32.4°C; (AAAC)7, (AAAG)7, (CATT)7, (GAAT)7, and (GATA)7 at 41.0°C; (AAAT)15 at 43.7°C; (AAC)10, (AAG)10, (AGT)10, and (GAT)10 at 46.1°C; (GACA)7, (GGAA)7, and (GGAT)7 at 51.3°C; (CA)15 and (CT)15 at 52.9°C; (ACG)10, (CAG)10, (CCT)10, and (GGT)10 at 59.7°C; and (CCG)10 at 73.4°C. The two remaining dinucleotide motifs, (AT)n and (GC)n, were not included in the experiment because of the difficulty of using self-complementary probes. The filters were subsequently exposed to PhosphorImager screens for 4–40 hr, and images were scanned from the screens using a PhosphorImager scanner (Molecular Dynamics). The pixel values above background level were calculated for all dots using ImageQuant software (Molecular Dynamics) with a gray-scale range of 2–100 and local background. The 12 di- and trinucleotide oligonucleotide probes were first hybridized to filters and analyzed, after which filters were stripped with boiling water, exposed overnight as described above to ensure that probe molecules had been removed efficiently, and then screened with the 9 tetranucleotide probes.
To determine the absolute number of (CA)nmicrosatellites in the chicken genome along with calibrating hybridization stringency with the minimum length of repeats detected, a filter with human and chicken DNA dilutions together with five plasmid (pUC19) preparations of clones containing different sized (CA)n repeats (n = 10, 12, 14, 16, or 18) was also prepared. Plasmid dilutions equivalent to 25,000, 12,500, and 5,000 CA copies were included. Hybridization conditions and method of detection were as described above. The number of (CA)n repeats in the avian and human genomes was estimated from the calibration of signal intensities against known plasmid copy numbers.
Database Searching
Searches for all possible avian mono-, di-, tri-, and tetranucleotide motifs at least 20 bp in length (no mismatches allowed) were carried out on the (nonmammalian vertebrate) VRT subsection of GenBank release 93.0, using the FASTA program of the GCG computer package. Sequences annotated as derived from microsatellite library screenings were excluded. Hits corresponding to the poly(A) tails of cDNA clones were also excluded as were mononucleotide stretches apparently originating from terminal transferase treatment. In cases when the same microsatellite locus was present more than once in the database, only a single match was recorded.
One hundred genomic and 100 cDNA GenBank chicken entries were selected randomly using the Entrez World Wide Web site (http://www3.ncbi.nlm.nih.gov/Entrez/) to derive an estimate of the relative proportions of noncoding, transcribed but not translated (UTR), and coding DNA in the database. Estimates of the total number of base pairs in the different categories were subsequently obtained by extrapolating figures from this selection. Regions from entries not annotated to a particular category were allocated proportionally to the different groups. By relating the number of microsatellites found in coding and noncoding DNA, respectively, to the estimated number of base pairs contained in GenBank, microsatellite frequencies in the two types of DNA were calculated. Totally, there were 1026 genomic sequences and 1907 cDNA sequences in the database.
Primed In Situ Labeling
Chicken metaphase chromosome spreads were obtained from bone marrow using standard methods (e.g., Christidis 1983). Briefly, bone marrow cells from chicken femur were incubated in RPMI-1640 (Sigma), containing 30% fetal calf serum and 0.1 mg/ml of colcemid for 1 hr, followed by routine hypotonic treatment and fixation. The labeling reaction was based on the study of Winterø et al. (1992) as follows: 50 μl of reaction mixture that contained 150 pmoles of CA10 primer, 1 mm dNTP, 10 nm spectrum orange dUTP (Vysis Inc.), 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 2.0 mm MgCl2, and 5 units of AmpliTaq DNA polymerase (Perkin Elmer), was added to a metaphase slide, and a coverslip was added and sealed with rubber cement to limit evaporation. Slides were then incubated at 95°C for 5 min followed by a 1 hr incubation at 60°C or 65°C. The reaction was terminated by washing once in 50 mm NaCl, 50 mm EDTA at 60°C for 5 min followed by two 5 min washes at room temperature in 4× SSC, 0.1% Tween 20. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) that produced a G-band like pattern for chromosome identification. The results were analyzed using an Olympus BX60 fluorescence microscope equipped with an IMAC-CCD S30 video camera. Images were captured and analyzed using ISIS 1.65 (Metasystems) software.
Acknowledgments
We thank Fredrik Söderbom for assistance with the PhosphorImager analysis and Hugh Williams for assistance with database searching.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵3 Corresponding author.
-
E-MAIL Hans.Ellegren{at}bmc.uu.se; FAX +46-18-504461.
-
- Received December 18, 1996.
- Accepted March 3, 1997.
- Cold Spring Harbor Laboratory Press