variation graph data structures, interchange formats, alignment, genotyping, and variant calling methods
Variation graphs provide a succinct encoding of the sequences of many genomes. A variation graph (in particular as implemented in vg) is composed of:
- nodes, which are labeled by sequences and ids
- edges, which connect two nodes via either of their respective ends
- paths, describe genomes, sequence alignments, and annotations (such as gene models and transcripts) as walks through nodes connected by edges
This model is similar to sequence graphs that have been used in assembly and multiple sequence alignment.
Paths provide coordinate systems relative to genomes encoded in the graph, allowing stable mappings to be produced even if the structure of the graph is changed. The variation graph model makes this embedding explicit and essential. Tools in vg maintain paths as immutable during transformations of the graph. They use paths to project graph-relative data into reference-relative coordinate spaces. Paths provide stable coordinates for graphs built in different ways from the same input sequences.
We maintain a support forum on biostars: https://www.biostars.org/tag/vg/
The easiest way to get vg is to download one of our release builds for Linux. We have a 6-week release cadence, so our builds are never too far out of date.

Download the latest vg release for Linux
For MacOS, see Building on MacOS.
If you don't want to or can't use a pre-built release of vg, or if you want to become a vg developer, you can build it from source instead.
First, obtain the repo and its submodules:
git clone --recursive https://github.com/vgteam/vg.git
cd vg
Then, install VG's dependencies. You'll need the protobuf and jansson development libraries installed, and to run the tests you will need:
* jq, bc, rs, and parallel
* hexdump and column from bsdmainutils
* npm for testing documentation examples).
On Ubuntu, you should be able to do:
make get-deps
On other distros, you will need to perform the equivalent of:
sudo apt-get install build-essential git cmake pkg-config libncurses-dev libbz2-dev \
protobuf-compiler libprotoc-dev libprotobuf-dev libjansson-dev \
automake gettext autopoint libtool jq bsdmainutils bc rs parallel \
npm curl unzip redland-utils librdf-dev bison flex gawk lzma-dev \
liblzma-dev liblz4-dev libffi-dev libcairo-dev libboost-all-dev \
libzstd-devel pybind11-dev python3-pybind11
Note that Ubuntu 16.04 does not ship a sufficiently new Protobuf; vg requires Protobuf 3 which will have to be manually installed.
At present, you will need GCC version 4.9 or greater, with support for C++14, to compile vg. (Check your version with gcc --version.) GCC up to 11.2.0 is supported.
Other libraries may be required. Please report any build difficulties.
Note that a 64-bit OS is required. Ubuntu 20.04 should work.
When you are ready, build with . ./source_me.sh && make, and run with ./bin/vg.
Note that vg can take anywhere from 10 minutes to more than an hour to compile depending on your machine and the number of threads used.
You can also produce a static binary with make static, assuming you have static versions of all the dependencies installed on your system.
The first step is to clone the vg repository:
git clone --recursive https://github.com/vgteam/vg.git
cd vg
VG depends on a number of packages being installed on the system where it is being built. Dependencies can be installed using either MacPorts or Homebrew.
You can use MacPorts to install VG's dependencies:
sudo port install libtool protobuf3-cpp jansson jq cmake pkgconfig autoconf automake libtool coreutils samtools redland bison gperftools md5sha1sum rasqal gmake autogen cairo libomp boost zstd pybind11
Homebrew provides another package management solution for OSX, and may be preferable to some users over MacPorts. VG ships a Brewfile describing its Homebrew dependencies, so from the root vg directory, you can install dependencies, and expose them to vg, like this:
# Install all the dependencies in the Brewfile
brew bundle
With dependencies installed, VG can now be built:
. ./source_me.sh && make
Note that static binaries cannot yet be built for Mac.
Our team has successfully built vg on Mac with GCC versions 4.9, 5.3, 6, 7, and 7.3, as well as Clang 9.0.
The Mac platform is moving to ARM, with Apple's M1, M1 Pro, M1 Max, and subsequent chip designs. The vg codebase supports ARM on Mac as well as on Linux. The normal installation instructions work on a factory-fresh ARM Mac.
However, it is easy to run into problems when migrating a working vg build environment or migrating Macports or Homebrew from x86_64 to ARM. The ARM machine can successfully run x86_64 tools installed via Macports or Homebrew on the old machine, but vg can only build properly on ARM if you are using ARM versions of the build tools, like make and CMake.
So, after migrating to an ARM Mac using e.g. Apple's migration tools:
- Uninstall Macports and its packages, if they were migrated from the old machine. Only an ARM Macports install can be used to provide dependencies for vg on ARM.
- Uninstall Homebrew and its packages, if they were migrated. Similarly, only an ARM Homebrew install will work.
- Reinstall one of Macports or Homebrew. Make sure to use the M1 or ARM version.
- Use the package manager you installed to install system dependencies of vg, such as CMake, as documented above.
- Clean vg with
make clean. This should remove all build artefacts. - Build vg again with
make.
If you still experience build problems after this, delete the whole checkout and check out the code again; make clean is not under CI test and is not always up to date with the rest of the build system.
Whether or not that helps, please then open an issue so we can help fix the build or fix make clean.
Note See the
vg autoindexexamples below for how to use that tool in place ofvg constructto build and index graphs in a single step.
One way to build a graph with vg is to construct it from variant calls using a reference FASTA file and VCF file. If you're working in vg's test/ directory:
vg construct -r small/x.fa -v small/x.vcf.gz >x.vgNote that to build a graph, an index of the VCF file is required. The VCF index file can be generated using the tabix command provided by SAMtools (e.g. tabix -p vcf x.vcf.gz on the command line).
You can also build a graph (and indexes for mapping with vg) from a set of genome assemblies (FASTA), as opposed to variant calls as described above, using Minigraph-Cactus.
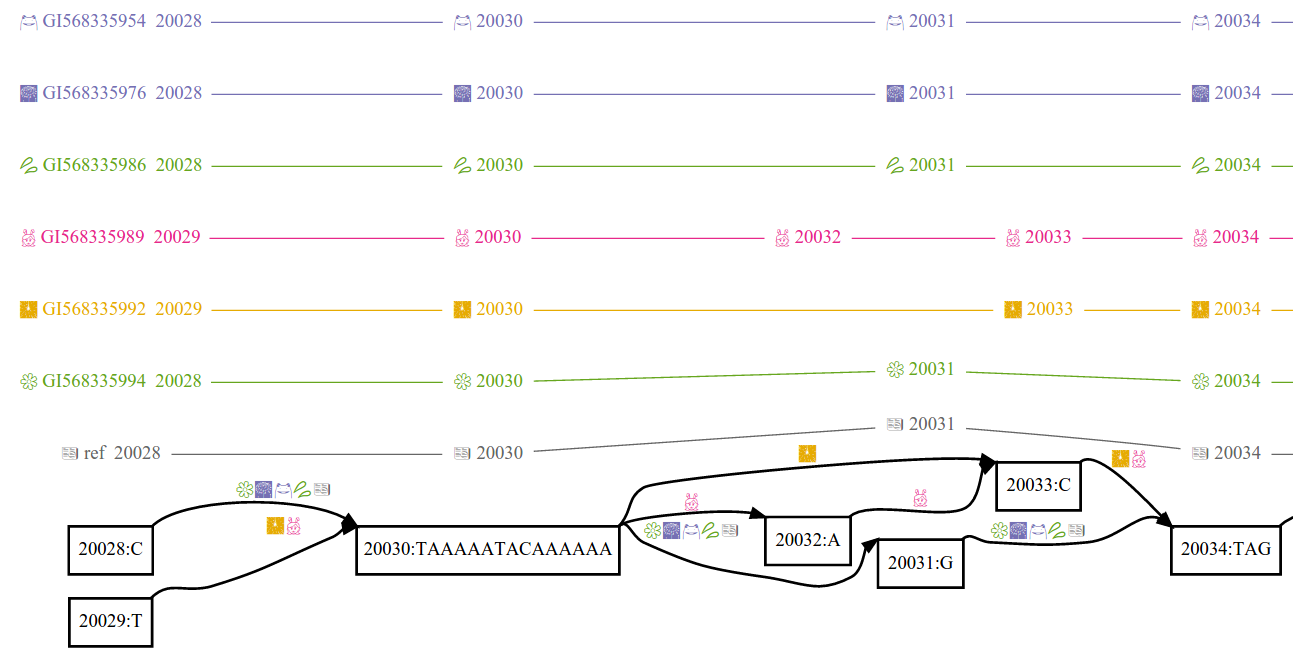
vg supports many formats, the three most important are:
PackedGraph (.vg): This isvg'snative format. It supports edits of all kinds (to topology and paths), but can be inefficient at large scales, especially with many paths.GFA (.gfa): GFA is standard text-based format and usually the best way to exchange graphs betweenvgand other pangenome tools.vgcan also operate on (uncompressed) GFA files directly, by way of using aPackedGraphrepresentation in memory (and therefore shares that format's scaling concerns and edit-ability).GBZ (.gbz): GBZ is a highly-compressed format that uses much less space to store paths than the above formats, but at the cost of not allowing general edits to the graph.
You can query the format of any graph using vg stats -F.
In general, you will build and index vg graphs using vg autoindex (from GFA or VCF) or Minigraph-Cactus (FASTAs). You can also import GFA files from other tools such as ODGI and PGGB using vg convert -g.
You can convert any graph to GFA using vg convert -f. By default, vg uses GFA v1.1 where paths are represented as W-lines. To use P-lines instead (GFA v1.0), use vg convert -fW.
The GBZ format makes the distinction between REFERENCE and HAPLOTYPE paths. REFERENCE paths can be used as coordinate systems but are more expensive to store. HAPLOTYPE paths are highly compressed but cannot be used for position lookups. In the HPRC graphs for example, contigs from GRCh38 and CHM13(T2T) are REFERENCE paths and all other samples HAPLOTYPE paths.
The distinction between REFERENCE and HAPLOTYPE paths is carried over into the other formats such as .vg and .gfa to facilitate conversion and inter-operation. In .gfa, REFERENCE paths are P-Lines, or W-lines whose sample names are flagged in the header. W-lines whose names are not flagged in the header are HAPLOTYPE paths. In .vg they are denoted using a naming convention.
See the Path Metadata WIKI for more details.
Warning
GBZis the only format that supports efficient loading large numbers ofHAPLOTYPEpaths invg. You may run into issues trying to load whole-genome graphs with thousands ofHAPLOTYPEfrom.vgor.gfafiles.vg convert -Hcan be used to dropHAPLOTYPEpaths, allowing the graph to be more easily loaded in other formats.
Note It is best to use the newer
vg converttool (described above) for GFA conversion
vg view provides a way to convert the graph into various formats:
# GFA output
vg view x.vg >x.gfa
# dot output suitable for graphviz
vg view -d x.vg >x.dot
# And if you have a GAM file
cp small/x-s1337-n1.gam x.gam
# json version of binary alignments
vg view -a x.gam >x.jsonIf you have more than one sequence, or you are working on a large graph, you will want to map rather than merely aligning.
There are multiple read mappers in vg:
vg giraffeis designed to be fast for highly accurate short reads, against graphs with haplotype information.vg mapis a general-purpose read mapper.vg mpmapdoes "munti-path" mapping, to allow describing local alignment uncertainty. This is useful for transcriptomics.
To use vg giraffe to map reads, you will first need to prepare indexes. This is best done using vg autoindex. In order to get vg autoindex to use haplotype information from a VCF file, you can give it the VCF and the associated linear reference directly.
# construct the graph and indexes (paths below assume running from `vg/test` directory)
vg autoindex --workflow giraffe -r small/x.fa -v small/x.vcf.gz -p x
# simulate a bunch of 150bp reads from the graph, into a GAM file of reads aligned to a graph
vg sim -n 1000 -l 150 -x x.giraffe.gbz -a > x.sim.gam
# now re-map these reads against the graph, and get BAM output in linear space
# FASTQ input uses -f instead of -G.
vg giraffe -Z x.giraffe.gbz -G x.sim.gam -o BAM > aln.bamMore information on using vg girafe can be found on the vg wiki.
If your graph is large, you want to use vg index to store the graph and vg map to align reads. vg map implements a kmer based seed and extend alignment model that is similar to that used in aligners like novoalign or MOSAIK. First an on-disk index is built with vg index which includes the graph itself and kmers of a particular size. When mapping, any kmer size shorter than that used in the index can be employed, and by default the mapper will decrease the kmer size to increase sensitivity when alignment at a particular k fails.
# construct the graph (paths below assume running from `vg/test` directory)
vg construct -r small/x.fa -v small/x.vcf.gz > x.vg
# store the graph in the xg/gcsa index pair
vg index -x x.xg -g x.gcsa -k 16 x.vg
# align a read to the indexed version of the graph
# note that the graph file is not opened, but x.vg.index is assumed
vg map -s CTACTGACAGCAGAAGTTTGCTGTGAAGATTAAATTAGGTGATGCTTG -x x.xg -g x.gcsa > read.gam
# simulate a bunch of 150bp reads from the graph, one per line
vg sim -n 1000 -l 150 -x x.xg > x.sim.txt
# now map these reads against the graph to get a GAM
vg map -T x.sim.txt -x x.xg -g x.gcsa > aln.gam
# surject the alignments back into the reference space of sequence "x", yielding a BAM file
vg surject -x x.xg -b aln.gam > aln.bam
# or alternatively, surject them to BAM in the call to map
vg sim -n 1000 -l 150 -x x.xg > x.sim.txt
vg map -T x.sim.txt -x x.xg -g x.gcsa --surject-to bam > aln.bamVariation from alignments can be embedded back into the graph. This process is called augmentation and can be used for de novo variant calling, for example (see below).
Warning Using
vg augmentfor variant calling remains very experimental. It is not at all recommended for structural variant calling, and even for small variants, you will often get much more accurate results (at least on human) by projecting your alignment toBAMand running a linear variant caller such as DeepVariant.
# augment the graph with all variation from the GAM except that implied by soft clips, saving to aug.vg. aug.gam contains the same reads as aln.gam but mapped to aug.vg
vg augment x.vg aln.gam -A aug.gam > aug.vg
# augment the graph with all variation from the GAM, saving each mapping as a path in the graph.
# softclips of alignment paths are preserved (`-S`).
# Note, this can be much less efficient than the above example if there are many alignments in the GAM
vg augment x.vg aln.gam -i -S > aug_with_paths.vgNote More information can be found in the WIKI.
The following examples show how to generate a VCF with vg using read support. They depend on output from the Mapping and Augmentation examples above. Small variants and SVs can be called using the same approach. Currently, it is more accuracte for SVs.
Call only variants that are present in the graph:
# Compute the read support from the gam
# -Q 5: ignore mapping and base qualitiy < 5
vg pack -x x.xg -g aln.gam -Q 5 -o aln.pack
# Generate a VCF from the support.
vg call x.xg -k aln.pack > graph_calls.vcfBy default, vg call omits 0/0 variants and tries to normalize alleles to make the VCF more compact. Both these steps can make it difficult to compare the outputs from different samples as the VCFs will have different coordinates even though they were created using the same graph. The -a option addresses this by calling every snarl using the same coordinates and including reference calls. Outputs for different samples can be combined with bcftools merge -m all.
vg call x.xg -k aln.pack -a > snarl_genotypes.vcf
In order to also consider novel variants from the reads, use the augmented graph and gam (as created in the "Augmentation" example using vg augment -A):
Warning Using
vg augmentfor variant calling remains very experimental. It is not at all recommended for structural variant calling, and even for small variants, you will often get much more accurate results (at least on human) by projecting your alignment toBAMand running a linear variant caller such as DeepVariant.
# Index our augmented graph
vg index aug.vg -x aug.xg
# Compute the read support from the augmented gam (ignoring qualitiy < 5, and 1st and last 5bp of each read)
vg pack -x aug.xg -g aug.gam -Q 5 -s 5 -o aln_aug.pack
# Generate a VCF from the support
vg call aug.xg -k aln_aug.pack > calls.vcfA similar process can by used to genotype known variants from a VCF. To do this, the graph must be constructed from the VCF with vg construct -a (graphs from other sources such as vg autoindex and Minigraph-Cactus cannot be used):
# Re-construct the same graph as before but with `-a`
vg construct -r small/x.fa -v small/x.vcf.gz -a > xa.vg
# Index the graph with `-L' to preserve alt paths in the xg
vg index xa.vg -x xa.xg -L
# Compute the support (we could also reuse aln.pack from above)
vg pack -x xa.xg -g aln.gam -o aln.pack
# Genotype the VCF (use -v)
vg call xa.xg -k aln.pack -v small/x.vcf.gz > genotypes.vcfPre-filtering the GAM before computing support can improve precision of SNP calling:
# filter secondary and ambiguous read mappings out of the gam
vg filter aln.gam -r 0.90 -fu -m 1 -q 15 -D 999 -x x.xg > aln.filtered.gam
# then compute the support from aln.filtered.gam instead of aln.gam in above etc.
vg pack -x xa.xg -g aln.filtered.gam -o aln.pack
vg call xa.xg -k aln.pack -v small/x.vcf.gz > genotypes.vcfFor larger graphs, it is recommended to compute snarls separately:
vg snarls x.xg > x.snarls
# load snarls from a file instead of computing on the fly
vg call x.xg -k aln.pack -r x.snarls > calls.vcfNote: vg augment, vg pack, vg call and vg snarls can now all be run on directly on any graph format (ex '.gbz', '.gfa', .vg, .xg (except augment) or anything output by vg convert). Operating on .vg or '.gfa' uses the most memory and is not recommended for large graphs. The output of vg pack can only be read in conjunction with the same graph used to create it, so vg pack x.vg -g aln.gam -o x.pack then vg call x.xg -k x.pack will not work.
Infer variants from from alignments implied by paths in the graph. This can be used, for example, to call SVs directly from a variation graph that was constructed from a multiple alignment of different assemblies:
# create a graph from a multiple alignment of HLA haplotypes (from vg/test directory)
vg msga -f GRCh38_alts/FASTA/HLA/V-352962.fa -t 1 -k 16 | vg mod -U 10 - | vg mod -c - > hla.vg
# index it
vg index hla.vg -x hla.xg
# generate a VCF using gi|568815592:29791752-29792749 as the reference contig. The other paths will be considered as haploid samples
vg deconstruct hla.xg -e -p "gi|568815592:29791752-29792749" > hla_variants.vcfVariants can also be inferred strictly from topology by not using -e, though unlike the above example, cycles are not supported. "Deconstruct" the VCF variants that were used to construct the graph. The output will be similar but identical to small/x.vcf.gz as vg construct can add edges between adjacent alts and/or do some normalization:
# using the same graph from the `map` example
vg deconstruct x.xg -p x > x.vcfHaplotype paths from .gbz or .gbwt indexes input can be considered using -z and `-g', respectively.
As with vg call, it is best to compute snarls separately and pass them in with -r when working with large graphs.
vg has a number of tools to support transcriptomic analyses with spliced graphs (i.e. graphs that have annotated splice junctions added as edges into the graph). These edges can be added into an existing graph using vg rna. We can then perform splice-aware mapping to these graphs using vg mpmap. vg developers have also made a tool for haplotype-aware transcript quantification based on these tools in rpvg. The easiest way to start this pipeline is to use the vg autoindex subcommand to make indexes for vg mpmap. vg autoindex creates indexes for mapping from common interchange formats like FASTA, VCF, and GTF.
More information is available in the wiki page on transcriptomics.
Working from the test/ directory the following example shows how to create a spliced pangenome graph and indexes using vg autoindex with 4 threads:
# Create spliced pangenome graph and indexes for vg mpmap
vg autoindex --workflow mpmap -t 4 --prefix vg_rna --ref-fasta small/x.fa --vcf small/x.vcf.gz --tx-gff small/x.gtfRNA-seq reads can be mapped to the spliced pangenome graph using vg mpmap with 4 threads:
# Map simulated RNA-seq reads using vg mpmap
vg mpmap -n rna -t 4 -x vg_rna.spliced.xg -g vg_rna.spliced.gcsa -d vg_rna.spliced.dist -f small/x_rna_1.fq -f small/x_rna_2.fq > mpmap.gampThis will produce alignments in the multipath format. For more information on the multipath alignment format and vg mpmap see wiki page on mpmap. Running the two commands on the small example data using 4 threads should on most machines take less than a minute.
If you have a small graph, you can align a sequence to the whole graph, using a full-length partial order alignment:
vg align -s CTACTGACAGCAGAAGTTTGCTGTGAAGATTAAATTAGGTGATGCTTG x.vgNote that you don't have to store the graph on disk at all, you can simply pipe it into the local aligner:
vg construct -r small/x.fa -v small/x.vcf.gz | vg align -s CTACTGACAGCAGAAGTTTGCTGTGAAGATTAAATTAGGTGATGCTTG -Most commands allow the streaming of graphs into and out of vg.
A variety of commands are available:
- autoindex: construct graphs and indexes for other tools from common interchange file formats
- construct: graph construction
- index: index features of the graph in a disk-backed key/value store
- map: mapp reads to a graph
- giraffe: fast, haplotype-based mapping of reads to a graph
- mpmap: short read mapping and multipath alignment (optionally spliced)
- surject: project graph alignments onto a linear reference
- augment: adds variation from aligned reads into the graph
- call: call variants from an augmented graph
- rna: construct splicing graphs and pantranscriptomes
- convert: convert graph and alignment formats
- combine: combine graphs
- chunk: extract or break into subgraphs
- ids: node ID manipulation
- sim: simulate reads by walking paths in the graph
- prune: prune graphs to restrict their path complexity
- snarls: find bubble-like motifs in a graph
- mod: various graph transformations
- filter: filter reads out of an alignment
- deconstruct: create a VCF from variation in the graph
- paths: traverse paths in the graph
- stats: metrics describing graph properties
vg is a collection of tools based on a common data model (the variation graph) that is described by a protobuf schema (vg.proto). Data objects defined in vg.proto may be serialized via a stream pattern defined in stream.hpp. It is not necessary to write code in vg in order to interface with the algorithms defined here. Rather, it is sometimes simpler to write an external algorithm that reads and writes the same data formats.
MIT