Abstract
Image quality control (QC) is a critical and computationally intensive component of functional magnetic resonance imaging (fMRI). Artifacts caused by physiologic signals or hardware malfunctions are usually identified and removed during data processing offline, well after scanning sessions are complete. A system with the computational efficiency to identify and remove artifacts during image acquisition would permit rapid adjustment of protocols as issues arise during experiments. To improve the speed and accuracy of QC and functional image correction, we developed Fast Anatomy-Based Image Correction (Fast ANATICOR) with newly implemented nuisance models and an improved pipeline. We validated its performance on a dataset consisting of normal scans and scans containing known hardware-driven artifacts. Fast ANATICOR’s increased processing speed may make real-time QC and image correction feasible as compared with the existing offline method.
Keywords: Functional MRI, Real-Time fMRI, Resting-State Connectivity, Sliding-Windowed Timeseries, Online Denoising, Artifact Detection
1. Introduction
Time is a significant experimental consideration in fMRI studies for both of image quality monitoring and image data processing because one scanning session can create scores of discrete images at a repetition time (TR) around 2-4 s for conventional scanning protocols. Comprehensive image quality control (QC) measures and nuisance models encompassing multiple sources of artifacts are also essential to effective experiments, but it is computationally time consuming, and necessitating QC processing offline when acquisition is finished.
Multiple distinct artifacts impact the quality of echo planar imaging (EPI) time series data. Head motion, physiologic interference (cardiac pulse and respiration), and hardware artifacts [1-4] can all be sources of error [5], and can be missed without a comprehensive approach to their identification [3]. Denoising methods have primarily focused on head motion and physiological inferences [6, 7], and head motion has been the only QC measure that can be identified during scanning [8], leaving hardware artifacts to be identified afterwards. Not all hardware artifacts are apparent upon visual inspection of standard EPI time series, but a hardware malfunction that has corrupted a dataset may be identified in correlation maps during image analysis, after multiple subjects have been scanned under non-ideal conditions [3]. Therefore, the various applications of fMRI would be best served by a monitoring system capable of data quality control during scanning and subsequently permitting rapid adjustments of experimental protocols [5, 9-11].
Here, we introduce a new model named Fast Anatomy-Based Image Correction (Fast ANATICOR) that can monitor image quality and detect and remove transient local artifacts within relatively shorter time than the traditional full-offline methods. Improving upon the anatomy-based image correction (ANATICOR), Fast ANATICOR uses nuisance derived signals, new metrics, and a new pipeline to demonstrate remarkably enhanced processing efficiency as compared with traditional approaches. In this model, we use a weighted white matter (WM) regressor to focus on a local transient artifact as the unique signature of coil instability or time series defects; artifacts which may be visually undetectable in EPI data and difficult to detect using existing QC metrics. This improved and experimentally validated denoising approach permits the removal of artifacts from EPI data during image acquisition. Based on our investigation, we further propose a denoising method for sliding-time-windowed (30 TRs) connectivity applications [12-14].
2. Methods
2.1. Subjects
Fifteen right-handed males (mean age 20±2.14 years) without a known history of neurological or psychiatric disorders participated in this study. Written, informed assent and consent were obtained from all participants and/or their parent/guardian. The National Institutes of Health Institutional Review Board approved the study.
2.2. Image acquisition
Subjects were sequentially scanned using an 8-channel head coil array equipped GE 3 Tesla (3T) scanner. For six of the total fifteen scans, the scanner had a hardware system malfunction identified to be a defect in one channel of the head coil. Subjects N01-N09 comprised the normal data set and were scanned in the absence of the head coil defect. Subjects A01-A06 comprised the abnormal data set and were scanned using the same head coil, while the head coil defect was present. Subject order and scan dates are shown in Table 1. During scanning sessions, all subjects were instructed to fixate on a cross in the center of the screen. Resting-state (RS) fMRI time series were acquired for 490 s (140 volumes) by a T2*-weighted gradient echo pulse sequence with high spatial resolution (1.72×1.72×3.00 mm3, TR=3.5 s, TE=27 ms, flip angle = 90 °). Respiration volume and heart rhythm were monitored with a respiration belt and pulse oximeter. Belt diameter and pulse oximeter readings were sampled at 50 Hz during scanning. Two high-resolution (0.94×0.94×1.20 mm3) T1-weighted anatomical images were also acquired with an MPRAGE sequence.
Table 1.
Data qualification measures.
| Subject Index |
Head Coil Artifact Detectable by |
Inter-TR Intensity Difference (%) |
Head Motion |
|||||
|---|---|---|---|---|---|---|---|---|
| WMeLOCAL | GCOR | CVAR (tNSR) | Original Time series |
Motion- corrected |
Outlier Volume |
Euclidean L2 norm (“enorm”) |
Outlier Volume |
|
| N01 | - | - | – | 0.44±0.29 | 0.68±1.11 | 0 | 0.02±0.01 | 0 |
| A01 | Y | N | N | 0.41±0.42 | 0.37±0.42 | 0 | 0.05±0.03 | 0 |
| N02 | - | - | - | 0.38±0.07 | 0.13±0.09 | 0 | 0.06±0.04 | 0 |
| N03 | - | - | - | 0.38±0.01 | 0.50±0.68 | 0 | 0.03±0.02 | 0 |
| A02 | Y | N | N | 0.32±0.04 | 0.10±0.10 | 0 | 0.02±0.01 | 0 |
| A03 | Y | N | N | 0.43±0.18 | 0.09±0.11 | 0 | 0.02±0.01 | 0 |
| N04 | - | - | - | 0.31±0.63 | 0.37±0.20 | 0 | 0.06±0.03 | 0 |
| N05 | - | - | - | 0.39±0.05 | 0.36±0.30 | 0 | 0.03±0.02 | 0 |
| N06 | - | - | - | 0.46±0.41 | 0.37±0.48 | 0 | 0.03±0.02 | 0 |
| N07 | - | - | - | 0.29±0.06 | 0.10±0.05 | 0 | 0.03±0.02 | 0 |
| A04 | Y | N | N | 0.34±0.09 | 0.32±0.27 | 0 | 0.03±0.02 | 0 |
| A05 | Y | N | N | 0.31±0.05 | 0.21±0.26 | 0 | 0.02±0.02 | 0 |
| N08 | - | - | - | 0.39±0.02 | 0.22±0.23 | 0 | 0.02±0.02 | 0 |
| A06 | Y | Y | N | 0.34±0.09 | 0.46±0.48 | 0 | 0.02±0.01 | 0 |
| N09* | - | - | - | 0.39±0.13 | 0.39±0.13 | 0 | 0.05±0.05 | 2 |
The subject indices were sequentially sorted by scan dates. Subject indices starting with “N” or “A” represent normal cases and artifactual cases where a hardware defect was present during scanning. WMeLOCAL, GCOR, CVAR, and tNSR represent local white matter signal, global correlation, coefficient of variance, and temporal noise-to-signal ratio, respectively. QC measures in this study were implemented in the AFNI package [18], fmriprep [25,26], and rtQC toolbox (DOI: 10.5281/zenodo.3239084).
The data sets from the 15 subjects were assessed by three QC tests assessing local white matter signal: local white matter regressor (WMeLOCAL) [3], global correlation (GCOR) [15], and coefficients of variance of EPI data (CVAREPI) [16] (see Table 1). The GCOR measure is computed as the brain-wide average correlation over all possible combinations of voxel timeseries. The calculation of GCOR is simplified by calculating the average dot product of the average unit-variance time series. The CVAR measure is computed the coefficient of variation of input voxels, which is the standard deviation of timeseries divided by the absolute value of mean timeseries.
2.3. Defining the target artifact
Unlike head motion artifacts, which can be visualized in EPI data during scanning, hardware artifacts may be revealed when correlation maps show marked biases that violate neurophysiological norms. To target a hardware artifact, one might consider a persistent or transient, local, abnormal correlation between brain regions. In a previous study, we found that the temporal signature of a local artifact could be found across the time series of multiple tissue voxels within a region imaged by a defective array component, not exclusively in deep WM regions that could have less partial volume effect with gray matter voxels [3]. Consequently, there should be high correlations between the time series in a voxel in the artifact region and in the time series in the surrounding area, assuming no involvement of other, stronger artifacts such as large head motion [17].
All procedures in this study used AFNI packages (http://afni.nimh.nih.gov) [18]. The high-resolution anatomical image was aligned to the fifth EPI volume of the EPI time series [19], and then processed with texture-based tissue classification to get the WM masks of individual subjects [20]. All WM masks were then resampled to EPI resolution. To reduce partial volume effects across different tissue types, we eroded WM masks by one voxel along each of the three axes. The first 4 volumes of the RS time series were removed to ensure that all remaining volumes in the time series were at magnetization steady state. Despiking was done to suppress the spiky artifact from rapid head motion and to enhance the accuracy of motion correction [17]. Rigid-body registration was used to estimate subject movement during RS EPI scans and to correct for slice timing [9]. All MRI images presented in this work were spatially normalized to the N27 brain template for ease of interpretation by brain region [21].
To precisely identify the target artifact, we began with a basic regression model for RS fMRI, defined as follows:
| (1:Model BASE) |
Here, yi is the EPI time series vector at voxel i. XMO is the matrix of second order Legendre polynomials to fit quadratic temporal trends and six regressors containing rigid-body motion parameter estimates (three translations and three rotations) intended to model any residual effects of movement after motion correction. XRI is the matrix of eight slice-wise regressors for retrospective image correction (RETROICOR) that models the effect of respiration and heart cycle [1], and a respiration volume per time (RVT) regressor that models slow blood oxygenation level fluctuations [2]. In Eq. (1) and subsequent equations, is the residual signal referring to the RS fMRI signal with unwanted nuisance components projected out.
The model for ANATICOR is defined in Eq. (2), where is the average local WM signal for each voxel i:
| (2:Model ANATICOR) |
Using the spatial pattern of correlation maps with the seed point at right middle frontal gyrus (RMFG) and marginal explained variance (R2) maps for the regressor , the subjects were categorized into two groups; normal cases (N01-N09) and cases with hardware artifacts (A01-A06).
2.4. Outlier tests
We used QC measures in the AFNI package with three criteria: the outlier test, head motion estimation, and GCOR [15]. In the outlier test, at each time point of EPI data, the inter-TR image differences were calculated as the number of outlier time points. In each time series, time points that differed from the trend were deemed outliers. The threshold level for the outlier time point (ethr) was defined by:
| (Eq. 3) |
Where, N is the length of time series, M is the median absolute deviation of the time series, and D−1 is the inverse of the reversed Gaussian cumulative distribution function. For all subjects, the number of outlier voxels was less than 1 percent of the number of voxels in the brain mask, and the Euclidean L2-norms (“enorm” in the AFNI package) of head movement were less than 0.25 (a.u.). Therefore all subjects met a practical threshold for minimizing artificial signal bias by head motion [17]. Additionally, GCOR of the whole brain was calculated to detect a concentrated correlation pattern that might be a signature of artifactual time courses [15], with the temporal noise-to-signal-ratio (tNSR) of EPI time series as a typical QC measure.
2.5. Artifact removal for fast denoising
For fast denoising, the nuisance model utilizing WMeLOCAL (ANATICOR) has been limited by calculation time. For this data set, calculating the localized WM signal (WMeLOCAL) for a single volume took over fifty TRs on average. To secure fast processing within 1 TR time, we replaced the localized regressor with isotropic Gaussian-distribution weights by distance from the voxel i:
| (Eq. 4: Model Weighted ANATICOR) |
Where, Wi is a vector for the distance weight by isotropic Gaussian kernel of full-width-at-half-maximum (FWHM) of 30 mm at each voxel i. The processing time was remarkably reduced in this model, in part by replacing the voxel loop in the pipeline with a weighted white matter regressor (wWMeLOCAL). Diagrams of the kernel weighting methods are presented in Fig. 1.
Fig. 1.
Concept diagrams for kernel weighting methods by local white matter regressor (WMeLocal) and weighted local white matter regressor (wWMeLocal) for ANATICOR and Fast ANATICOR models, respectively. WMeLocal is obtained by averaging voxel time series with a binary mask (B2) that intersects the eroded WM mask (A) and a binary kernel weight mask with radius r (B1). wWMeLocal is computed by averaging voxel time series with the weighted masks (B2) overlapped with the eroded WM mask (A) over a Gaussian curve (C1). The wWMeLocal method blurs the time series of white matter voxels using a Gaussian kernel with the given full-width-at-half-maximum (FWHM).
The local WM regressor considers the strength of covarying artifacts from each head coil array, which will differ by distance from the voxel i (see Figs. 1-C1 and 1-C2).
We replaced XRI (9 components) with respiration volume per time XRVT (1 component) to decrease processing time and to secure degrees of freedom for residual time series at the 30-TR time window.
| (Eq. 5: Model Fast ANATICOR; FANATICOR) |
The models BASE, ANATICOR, Weighted ANATICOR, and Fast ANATICOR (FANATICOR) contain 18, 19, 19, and 11 regressors, respectively.
2.6. Artifact indicators for image quality control
To demonstrate the transient nature of the local artifact, we used the R2 and tNSR maps of the wWMeLOCAL regressor to visualize the spatial distribution of the hardware artifact over time. For time windows of 30 TRs, we calculated the tNSR for two seed points, one in the LMFG and one in the artifactual region, the RMFG. To evaluate fast QC in those windows, we directly compared R2 maps of the nuisance regressors used in the denoising regression models: NULL, MO, MO+RVT, and Fast ANATICOR.
2.7. Spatial patterns of captured artifact and seed-based correlation maps within the time window
To visually inspect the spatiotemporal characteristics of the transient artifact for the entire time-windowed series (105 sets of 30 TR time windows), we obtained the R2 maps of the nuisance regressor used in the denoising regression models (NULL, MO, MO+RVT, and Fast ANATICOR) for normal and abnormal scans. To determine if each nuisance model could appropriately remove the targeted transient artifact over the full time series, we obtained the seed-based correlation maps for the RMFG seed point within the artifactual region for the artifactual group. The correlation and R2 maps from the artifactual subjects were compared with those of the normal subjects.
2.8. Variances of sliding-time-windowed correlations
Typically, real-time fMRI applications consider regions-of-interest (ROI) instead of individual voxels in order to shorten computation time and improve ease of interpretation (see [22], 2011, for review). To determine ROIs for each subject, EPI data were first split into 105 sets by 30-TR time windows. The residual time series were then obtained by the preprocessing pipeline (see Fig. 2) with the four denoising models (NULL, MO, MO+RVT, and Fast ANATICOR) and averaged within 90 ROIs using the 90-fROIs atlas from Shirer and colleagues [23]. For each denoising model and each individual subject, 105 time-windowed Pearson correlation matrices (90×90 matrix size) were calculated, and then transformed to Fisher’s z scores. To observe variance changes in time-windowed correlations for the different denoising models, the CVAR of time-windowed correlation (CVARρ) was calculated.
Fig 2.

Suggested pipeline for the real time application of image quality control and denoising. Asterisked (*) blocks indicate the procedural goals of this study; (i) to reduce time to the local white matter (WM) regressor and resolve the aliasing effect in weighting, (ii) to add a local WM regressor to the denoising regression process, and (iii) to get effective measurements for image quality control (QC). The ultimate goal is to perform the in-session procedures within 1 TR time.
2.9. Non-stationarity of sliding-time-windowed correlations
We also calculated the non-stationarity of the correlation matrices along the time axis by the augmented Dickey-Fuller unit root test [24]. Non-stationarity varied between the different preprocessing methods, with Fast ANATICOR showing the most non-stationarity. For individual time-windowed correlation matrices from an individual subject, we obtained a binary matrix representing significant non-stationarity at a threshold p < 0.05 from the unit root test. A frequency (the number of subjects) map showing significant non-stationary correlations was then calculated for the 15 individual subjects with each denoising model.
3. Results
3.1. Time costs of denoising and image quality control
The total elapsed time for nuisance regression by the Fast ANATICOR model was about 3.8 s for a single central processing unit (CPU), which is a marked improvement upon the previous method requiring 161.9 s per CPU on average. The calculation of the wWMeLOCAL regressor in the Fast ANATICOR model was about 52 times faster than the typical WMeLOCAL calculation in the ANATICOR model (see Table 2 for details). For QC with the wWMeLOCAL regressor, mapping tNSRwWMe could be accomplished at about 1 s per CPU, but mapping R2 took much longer, 5.8 s for a single CPU, on average (see Table 2).
Table 2.
Elapsed time for main processes of real-time monitoring and denoising.
| Subject Index |
4-core CPU system |
32-core CPU system |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Computing loads | Elapsed time for ANATICOR (s) |
Elapsed time for Fast ANATICOR (s) |
Elapsed time for ANATICOR (s) |
Elapsed time for Fast ANATICOR (s) |
||||||
| Number of WM Voxels in EPI data |
Number of GM Voxels in EPI data |
Obtaining WMe regressor |
Obtaining wWMeLOCAL regressor |
Denoising EPI time series |
Calculating explained variance (R2) of wWMeLOCAL |
Obtaining WMe regressor |
Obtaining wWMeLOCAL regressor |
Denoising EPI time series |
Calculating R2 of wWMeLOCAL |
|
| N01 | 27,141 | 85,188 | 153.975 | 0.796 | 3.218 | 6.112 | 25.763 | 0.333 | 0.736 | 1.119 |
| A01 | 26,713 | 77,079 | 157.376 | 0.799 | 2.958 | 5.580 | 26.329 | 0.333 | 0.693 | 1.030 |
| N02 | 21,591 | 75,389 | 153.020 | 0.793 | 2.880 | 5.428 | 25.603 | 0.332 | 0.680 | 1.005 |
| N03 | 27,152 | 87,690 | 155.563 | 0.793 | 3.349 | 6.325 | 26.027 | 0.332 | 0.758 | 1.154 |
| A02 | 24,641 | 74,194 | 156.392 | 0.794 | 2.816 | 5.360 | 26.165 | 0.332 | 0.669 | 0.993 |
| A03 | 20,779 | 75,559 | 157.839 | 0.792 | 2.866 | 5.425 | 26.407 | 0.332 | 0.678 | 1.004 |
| N04 | 26,331 | 86,913 | 159.751 | 0.798 | 3.289 | 6.218 | 26.725 | 0.333 | 0.748 | 1.136 |
| N05 | 21,869 | 76,548 | 158.471 | 0.791 | 2.947 | 5.558 | 26.512 | 0.332 | 0.691 | 1.026 |
| N06 | 24,992 | 82,229 | 158.998 | 0.790 | 3.110 | 5.880 | 26.600 | 0.332 | 0.718 | 1.080 |
| N07 | 27,707 | 81,464 | 159.190 | 0.794 | 3.100 | 5.897 | 26.632 | 0.332 | 0.717 | 1.083 |
| A04 | 26,008 | 87,839 | 159.451 | 0.793 | 3.319 | 6.262 | 26.675 | 0.332 | 0.753 | 1.144 |
| A05 | 16,869 | 77,247 | 156.130 | 0.798 | 2.910 | 5.517 | 26.122 | 0.333 | 0.685 | 1.020 |
| N08 | 20,813 | 74,151 | 157.754 | 0.796 | 2.863 | 5.376 | 26.392 | 0.333 | 0.677 | 0.996 |
| A06 | 31,060 | 84,439 | 163.981 | 0.802 | 3.192 | 6.087 | 27.430 | 0.334 | 0.732 | 1.115 |
| N09 | 24,343 | 77,640 | 159.198 | 0.790 | 2.937 | 5.568 | 26.633 | 0.332 | 0.690 | 1.028 |
| Mean ± standard deviation | 24,533.93 ±3,577.47 | 80,237.93 ±5,097.34 | 157.806 ±2.645 | 0.795 ±0.004 | 3.050 ±0.185 | 5.773 ±0.353 | 26.401 ±0.441 | 0.332 ±0.001 | 0.708 ±0.031 | 1.062 ±0.059 |
A larger voxel number is associated with greater elapsed time. Two computing systems were used for this time critical and complex analysis, a 4-core central processing unit (CPU) system (4.0 GHz clock speed; 32-gigabyte DDR3 memory; PCI-E solid-state drive) and a 32-core CPU system (3.0 GHz clock speed; 128-gigabytes DDR4 memory; PCI-E solid-state drive). The unit of time is seconds. The time window length is 30 repetition times (TRs). The subject indices were sorted in ascending order of scan dates.
3.2. Transient artifacts detected by weighted local white matter regressor
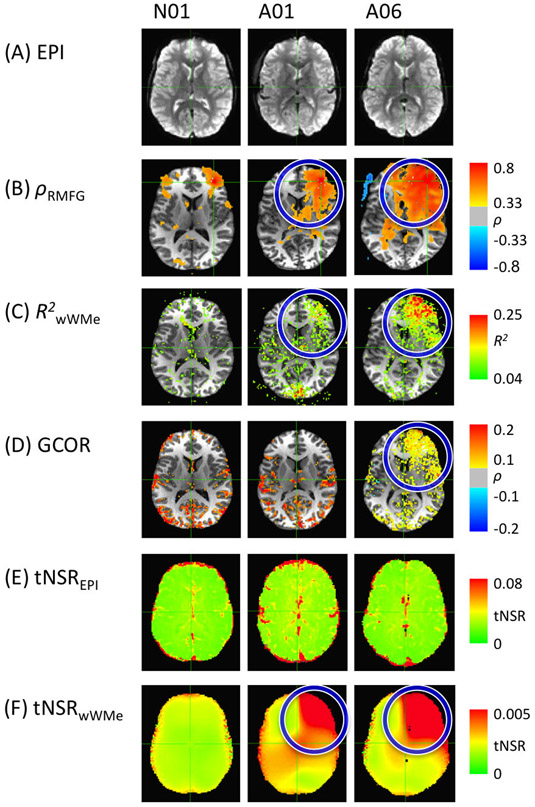
We tested the ability of nuisance models BASE, ANATICOR, Weighted ANATICOR, and of Fast ANATICOR to capture patterns of hardware artifacts present in our dataset. Fig. 3 shows how these indicators analyzed data from subject N01, who had a normal scan, and from subjects A01 and A06 whose scans had known hardware issues. In Fig. 3-A, there is no visual artifact pattern in the EPI data for both normal and abnormal scans, although abnormal scans did show abnormal correlation patterns between RMFG and other brain areas (see Fig. 3-B). Interestingly, we found that the tNSR of wWMeLOCAL and WMeLOCAL could detect the hardware artifact (see Fig. 3-F).
Fig. 3.
Approaches for transient artifact detection. In the echo planar imaging (EPI) data, (A) the artifactual pattern is not identifiable by visual inspection, (B) although the correlation maps could be biased by the artifact. (C) Local WM regressors can detect the artifact, but are time-consuming for high resolution data. (D) Global correlation (GCOR) measures only detect severely corrupted data. (E) Typical outlier detection or head motion estimation cannot detect a weak or partly corrupted artifact along the time axis. (F) Retrospective artifact detectors from deep WM structures can detect the artifactual pattern. Details are presented in the methods section.
In our dataset, the ANATICOR and Fast ANATICOR models differed significantly in processing time. The captured artifact patterns in R2 maps and in correlation maps with a seed point within the artifactual area (RMFG) were highly similar in the two models (see Fig. 4).
Fig. 4.
Correlation maps with a seed point at the right middle frontal gyrus ([31R, 45A, 10S] in the N27 brain template), which were derived from the residual time series after denoising regressions with different nuisance models, MO+RVT, ANATICOR, and Fast ANATICOR (FANATICOR). (A) For single subject data without coil artifact, there is no significant difference between correlation maps for all nuisance models (upper row), and a coil artifact pattern was not found in any of the explained variance (R2) maps of localized white matter (WM) regressor (lower row). (B) For single subject data with a significant coil artifact, a significantly biased correlation pattern was evident in and around the seed point for the MO+RVT nuisance model (first column, upper row). This coil artifact was corrected by the ANATICOR and Fast ANATICOR models (second and third columns, upper row). The artifacts were well captured by the localized WM regressor of the ANATICOR and Fast ANATICOR models (R2 maps, lower row).
The tNSR changes in wWMeLOCAL regressors (tNSRwWMe) at two seed locations within and outside of the coil artifact area, at the right and left middle frontal gyri (RMFG and LMFG), respectively, are presented in Fig. 5-A. When the transient artifact occurred in the RMFG, the tNSRwWMe at the RMFG increased. The tNSRwWMe at the LMFG, which was outside the artifactual area, did not change. During the time window when the artifact was absent [5, 34], the tNSRwWMe map showed no artifactual spatial patterns and the correlation maps for the RMFG seed calculated by the four denoising models were nearly indistinguishable (see Fig. 5-B) (see Fig. 5-B). Notably, during the time window [51, 80] when the artifact was present, the artifactual pattern was detected in the tNSRwWMe map, and the Fast ANATICOR model could clearly reduce its effect on the correlation map (see Fig. 5-C).
Fig. 5.
Spatiotemporal characteristics of transient artifacts detected by the weighted local white matter regressor (wWMeLOCAL), and performances of different denoising models for time-windowed correlations. (A) Temporal noise-to-signal ratio (tNSR) change in weighted local white matter regressors (wWMeLOCAL) at two seed locations within and outside of the coil artifact area are shown at the right and left middle frontal gyri (RMFG and LMFG), respectively. While the transient artifact occurs, tNSR is increased as shown by the red line. (B) For the time window [5, 34] TRs, the tNSR map of wWMeLOCAL (tNSRwWMe) and the correlation maps for the RMFG seed, which were calculated from 4 different denoising models, NONE, MO, MO+RVT, and Fast ANATICOR (FANATICOR) are shown. There is no significant coil artifact, no significant spatial pattern in the tNSRwWMe map, and no significant difference between the four correlation maps. (C) For the time window [51, 80] TRs, the tNSR map of wWMeLOCAL (tNSRwWMe) and the correlations between time series a seed point at RMFG and the residual time series from 4 different noise models (NONE, MO, MO+RVT, and Fast ANATICOR) are shown. This time interval contains a significant coil artifact reflected in its tNSRwWMe map. Only the Fast ANATICOR model could reduce the effect of the artifact.
3.3. Explained variances of nuisance regressors
We created R2 maps with a seed point in the RMFG for the normal and artifactual scans across the entire time series and compared them using each of the four denoising models. Fig. 6 shows this comparison for subject N01, who had no coil issues during scanning. As expected for the normal scan, no spatially concentrated signals were captured by the nuisance regressors of all the denoising models and the time-windowed correlation maps varied along time intervals with varying degrees and ranges in each model. Subjects N02-N09 showed similar results to N01.
Fig. 6.
For a subject with a normal scan (N01), changes in explained variance (R2) maps of artifact components from windowed time series for individual subjects were shown along with the correlation maps with a seed point at the right middle frontal gyrus. Changes in correlation maps along the time change shown in (A), (B1), (C1), and (D1) were denoised by the nuisance models NULL, MO, MO+RVT, and Fast ANATICOR (FANATICOR), respectively. The R2 maps of nuisance components are shown in (B2), (C2-C3), and (D2-D4) for each denoising models. The time window size is 30 TRs.
For the abnormal scan of subject A01, at time intervals with high tNSRwWMe values at the seed location, correlation patterns from the NULL, MO, and MO+RVT models showed the hardware artifact (see Figs. 7-A, B-1, and C-1), but R2 maps did not. Interestingly, R2 maps of wWMeLOCAL captured the hardware artifact at the appropriate time intervals (see Fig. 7-D4). The Fast ANATICOR model not only consistently identified the artifact, but effectively removed it, showing correlation patterns similar to those of the normal subjects at all time intervals (see Fig. 7-D1).
Fig. 7.
For a subject with an artifactual scan (A01), changes in explained variance (R2) maps of artifact components from windowed time series for individual subjects are shown along with the correlation maps with a seed point at right middle frontal gyrus. Changes in correlation maps along the time change shown in (A), (B1), (C1), and (D1), were denoised by the nuisance models NULL, MO, MO+RVT, and Fast ANATICOR (FANATICOR), respectively. The R2 maps of nuisance components are shown in (B2), (C2-C3), and (D2-D4) for each denoising model. The time window size is 30 TRs. As shown in the green circles in the rows (A), (B1), and (C1), the local transient artifacts occurred during the TR ranges [50, 79] and [75, 104]. Only the nuisance model Fast ANATICOR could detect those artifacts as shown in the green circle in the row D4, producing correlation maps (see row D1) comparable to the normal cases in Fig. 6.
3.4. Coefficients of variances for sliding-time-windowed correlations
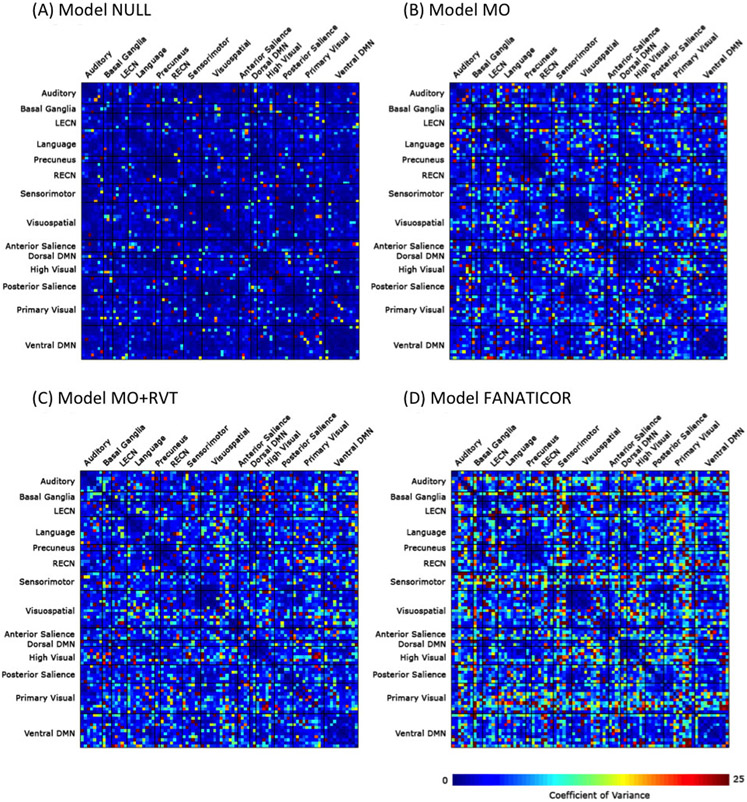
For all subjects and nuisance models used in this study, CVARρ matrices of time-windowed correlation matrices across the functional ROIs are shown in Fig. 8. The NULL and Fast ANATICOR models showed the fewest and most CVARρ values, respectively.
Fig. 8.
Coefficients of variance of windowed correlation (CVARρ) for different denoising models scaled to the 90 regions of interest (ROIs) of functional networks template. The NULL (panel A) and Fast ANATICOR (FANATICOR, panel D) models showed the least and most CVARρ values, respectively. The variances of time-windowed correlations along time axis for all functional ROIs were most increased by adding regressors.
3.5. Non-stationarity of sliding-time-windowed correlations
For all subjects and nuisance models in this study, the frequency matrices of subjects that showed significant non-stationary correlations are shown in Fig. 9. The NULL and Fast ANATICOR models showed the least and most non-stationarity, respectively. In general, the non-stationarity of time-windowed correlations was greater in correlations related to specific functional networks: auditory, basal ganglia, sensorimotor, visuospatial, anterior salience, and primary visual.
Fig. 9.
Non-stationarity of the correlation matrices along the time axis by the augmented Dickey-Fuller unit root test in the scale of the 90 regions of interest (ROIs) of functional networks template. For individual time-windowed correlation matrices from an individual subject, we obtained a binary matrix representing significant non-stationarity at a threshold p<0.05 from the unit root test. A frequency map of subjects that showed significantly non-stationary correlations was calculated from the 15 individual subjects. The NULL (panel A) and Fast ANATICOR (FANATICOR, panel D) models showed the least and most non-stationarity, respectively. An increase in the non-stationarity of time-windowed correlations was related to specific functional networks: auditory, basal ganglia, sensorimotor, visuospatial, anterior salience, and primary visual.
4. Discussion
4.1. Possibility of real-time denoising for sub-second repetition time
Calculation time limits the application of correction methods in real-time fMRI. The bottleneck created by the voxel-loop in ANATICOR’s pipeline has contributed significantly to processing time. We demonstrated marked temporal improvements to the ANATICOR model by replacing the voxel-loop with a weighted white matter regressor and using tNSR as an QC measure, thereby creating Fast ANATICOR (see Table 2, Fig. S1). With calculation speeds that permit QC during image acquisition, real-time implementation of Fast ANATICOR will depend on the processing stream and data transfer speeds. Sub-second processing for denoising can be accomplished by multi-core computing systems containing more than 12 computing cores or CPUs with parallel computing frameworks such as OpenMP libraries (http://openmp.org). Regarding system integration for real-time fMRI applications, however, more computing power might be required to operate accurate QC by tNSRwWMe, with real-time denoising by Fast ANATICOR. Current data transferring methods may need improvement in order to permit simultaneous transmission of data to both QC and denoising/analyzing systems that have been developed separately. Our suggested pipeline for real-time imaging applications is described in Fig. 2.
4.2. Variance and non-stationarity change in time-windowed correlations by denoising models
As more nuisance regressors were removed in the interest of calculation time, the CVARρ and non-stationarity in time-windowed correlations increased within the ROIs (Figs. 8 and 9). This trend was more evident in the Fast ANATICOR model than the MO or MO+RVT models, even though the head-coil artifact partially overlapped with the visuospatial, right executive control, and anterior salience networks. It could be suggested that nuisance signals operate like a drift process in the time course of time-windowed correlations and that the localized artifact could be a strong inducer of stationarity between the time-windowed correlations.
4.3. Fast ANATICOR and data quality
In this study, we used relatively high-resolution EPI data. Notably, the WM erosion step typically requires EPI data of a resolution comparable to or higher than that of the data used here. At lower-resolutions, such as 3.5 mm isocubic voxel, little remains of WM voxels after the erosion operation. Fast ANATICOR uses a Gaussian-curved weighting kernel to compute each voxel-wise regressor, giving more weight to the white matter that is closest to a given voxel. If there is no WM voxel close to a given voxel (due to a poor segmentation or stringent erosion operation in a low-resolution data grid, for example), the Gaussian curve will likely find WM voxels farther away instead of creating an empty regressor.
4.4. Physiological nuisance regressors
One physiological regressor, RVT, was included in this Fast ANATICOR model due to processing time limitations. In an interesting contrast to previous studies using typical, longer series of EPI data [3], the MO+RVT model in this study suppressed more artifactual correlations than NULL and MO models during the artifactual time interval (see Fig. 5-C). It is possible that physiological nuisance signals have a stronger effect in the input data with smaller time points in a time window analysis than the full-length EPI data.
5. Conclusion
In this study, we introduced a new image quality monitoring metric and denoising pipeline (Fast ANATICOR), optimized for a remarkably shorter processing time than traditionally ANATICOR. Using datasets with and without specific hardware artifacts, we validated Fast ANATICOR and compared it with other denoising models in terms of calculation time, artifact identification, and artifact removal. By optimizing the offline method with a weighted white matter regressor and streamlining the physiological regressors, we could both improve the detection of hardware artifacts and could decrease processing time.
Supplementary Material
Highlights:
Fast ANATICOR, a denoising method for real-time fMRI scanning, was developed to consider noise models for head motion, physiological, and non-physiological artifacts in image quality control.
Fast ANATICOR improved the detection of hardware artifacts and decreased processing time to within one repetition time (2-4s).
A weighted, local white matter signal proved an efficient and effective real-time image quality control metric.
The spatiotemporal characteristics of artifacts were reported along the time axis, demonstrating non-stationarity changes in sliding-time-window-based functional connectivity.
Acknowledgments
This work was supported by NIMH and NINDS Intramural Research Programs, a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI19C0218), and the Collaborative Genome Program for Fostering New Post-Genome Industry of the National Research Foundation (NRF) funded by the Ministry of Science and ICT (MSIT) (NRF-2017M3C9A6047623).
References
- [1].Glover GH, Li TQ, Ress D, Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR, Magn Reson Med, 44 (2000) 162–167. [DOI] [PubMed] [Google Scholar]
- [2].Birn RM, Diamond JB, Smith MA, Bandettini PA, Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI, Neuroimage, 31 (2006) 1536–1548. [DOI] [PubMed] [Google Scholar]
- [3].Jo HJ, Saad ZS, Simmons WK, Milbury LA, Cox RW, Mapping sources of correlation in resting state FMRI, with artifact detection and removal, Neuroimage, 52 (2010) 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Weiskopf N, Klose U, Birbaumer N, Mathiak K, Single-shot compensation of image distortions and BOLD contrast optimization using multi-echo EPI for real-time fMRI, Neuroimage, 24 (2005) 1068–1079. [DOI] [PubMed] [Google Scholar]
- [5].Voyvodic JT, Real-time fMRI paradigm control, physiology, and behavior combined with near real-time statistical analysis, Neuroimage, 10 (1999) 91–106. [DOI] [PubMed] [Google Scholar]
- [6].Mathiak K, Posse S, Evaluation of motion and realignment for functional magnetic resonance imaging in real time, Magn Reson Med, 45 (2001) 167–171. [DOI] [PubMed] [Google Scholar]
- [7].Misaki M, Barzigar N, Zotev V, Phillips R, Cheng S, Bodurka J, Real-time fMRI processing with physiological noise correction - Comparison with off-line analysis, J Neurosci Methods, 256 (2015) 117–121. [DOI] [PubMed] [Google Scholar]
- [8].Yang S, Ross TJ, Zhang Y, Stein EA, Yang Y, Head motion suppression using real-time feedback of motion information and its effects on task performance in fMRI, Neuroimage, 27 (2005) 153–162. [DOI] [PubMed] [Google Scholar]
- [9].Cox RW, Jesmanowicz A, Real-time 3D image registration for functional MRI, Magn Reson Med, 42 (1999) 1014–1018. [DOI] [PubMed] [Google Scholar]
- [10].Cox RW, Jesmanowicz A, Hyde JS, Real-time functional magnetic resonance imaging, Magn Reson Med, 33 (1995) 230–236. [DOI] [PubMed] [Google Scholar]
- [11].Cohen MS, Real-time functional magnetic resonance imaging, Methods, 25 (2001) 201–220. [DOI] [PubMed] [Google Scholar]
- [12].Chang C, Glover GH, Time-frequency dynamics of resting-state brain connectivity measured with fMRI, Neuroimage, 50 (2010) 81–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Handwerker DA, Roopchansingh V, Gonzalez-Castillo J, Bandettini PA, Periodic changes in fMRI connectivity, Neuroimage, 63 (2012) 1712–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ramot M, Kimmich S, Gonzalez-Castillo J, Roopchansingh V, Popal H, White E, Gotts SJ, Martin A, Direct modulation of aberrant brain network connectivity through real-time NeuroFeedback, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Saad ZS, Reynolds RC, Jo HJ, Gotts SJ, Chen G, Martin A, Cox RW, Correcting brain-wide correlation differences in resting-state FMRI, Brain Connect., 3 (2013) 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Handwerker DA, Gonzalez-Castillo J, D'Esposito M, Bandettini PA, The continuing challenge of understanding and modeling hemodynamic variation in fMRI, Neuroimage, 62 (2012) 1017–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jo HJ, Gotts SJ, Reynolds RC, Bandettini PA, Martin A, Cox RW, Saad ZS, Effective Preprocessing Procedures Virtually Eliminate Distance-Dependent Motion Artifacts in Resting State FMRI, Journal of Applied Mathematics, 2013 (2013) 935154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cox RW, AFNI: what a long strange trip it's been, Neuroimage, 62 (2012) 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Saad ZS, Glen DR, Chen G, Beauchamp MS, Desai R, Cox RW, A new method for improving functional-to-structural MRI alignment using local Pearson correlation, Neuroimage, 44 (2009) 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vovk A, Cox RW, Stare J, Suput D, Saad ZS, Segmentation priors from local image properties: without using bias field correction, location-based templates, or registration, Neuroimage, 55 (2011) 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC, Enhancement of MR images using registration for signal averaging, J Comput Assist Tomogr, 22 (1998) 324–333. [DOI] [PubMed] [Google Scholar]
- [22].LaConte SM, Decoding fMRI brain states in real-time, Neuroimage, 56 (2011) 440–454. [DOI] [PubMed] [Google Scholar]
- [23].Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD, Decoding subject-driven cognitive states with whole-brain connectivity patterns, Cereb Cortex, 22 (2012) 158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dickey DA, Fuller WA, Distribution of the Estimators for Autoregressive Time-Series with a Unit Root, J Am Stat Assoc, 74 (1979) 427–431. [Google Scholar]
- [25].Ciric R, Rosen AFG, Erus G, Cieslak M, Adebimpe A, Cook PA, Bassett DS, Davatzikos C, Wolf DH, Satterthwaite TD, Mitigating head motion artifact in functional connectivity MRI, Nat Protoc, 13 (2018) 2801–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Esteban O, Markiewicz CJ, Blair RW, Moodie CA, Isik AI, Erramuzpe A, Kent JD, Goncalves M, DuPre E, Snyder M, Oya H, Ghosh SS, Wright J, Durnez J, Poldrack RA, Gorgolewski KJ, fMRIPrep: a robust preprocessing pipeline for functional MRI, Nature methods, 16 (2019) 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.