Abstract
Genome-wide DNA methylation studies have quickly expanded due to advances in next-generation sequencing techniques along with a wealth of computational tools to analyze the data. Most of our knowledge about DNA methylation profiles, epigenetic heritability and the function of DNA methylation in plants derives from the model species Arabidopsis thaliana. There are increasingly many studies on DNA methylation in plants—uncovering methylation profiles and explaining variations in different plant tissues. Additionally, DNA methylation comparisons of different plant tissue types and dynamics during development processes are only slowly emerging but are crucial for understanding developmental and regulatory decisions. Translating this knowledge from plant model species to commercial crops could allow the establishment of new varieties with increased stress resilience and improved yield. In this review, we provide an overview of the most commonly applied bioinformatics tools for the analysis of DNA methylation data (particularly bisulfite sequencing data). The performances of a selection of the tools are analyzed for computational time and agreement in predicted methylated sites for A. thaliana, which has a smaller genome compared to the hexaploid bread wheat. The performance of the tools was benchmarked on five plant genomes. We give examples of applications of DNA methylation data analysis in crops (with a focus on cereals) and an outlook for future developments for DNA methylation status manipulations and data integration.
Keywords: epigenomics, epigenetics, bisulfite sequencing, DNA methylation, plants, differentially methylated regions
Introduction
Methylation of cytosine at carbon position 5 (also termed 5-meC) is a hallmark of an epigenetic modification, and 5-meC has been described as the fifth base of DNA [1]. Although the extent and context of 5-meC vary considerably between different plant lineages, all plants whose genomes have been sequenced and analyzed so far show substantial DNA methylation [2, 3]. Two major genomic contexts can be distinguished: (i) methylation on gene bodies and (ii) methylation on repeat sequences and transposons. Gene body methylation typically peaks on exons of moderately transcribed genes and, despite a comprehensive body of publications [3–5], its function remains mysterious [6]. Methylation on repeat sequences and transposons is crucial for suppressing transcription and is necessary for establishing heterochromatic domains. Consequently, mutations that abolish most DNA methylation lead to transposon activation and genomic meltdown after several generations in Arabidopsis thaliana. However, in early generations, the mutation can be outcrossed, and selfed offspring will be isogenic but with different DNA methylation states [7–9]. Experiments along these lines have established that these differences in DNA methylation can be stably inherited over many generations and influence ecologically relevant phenotypic traits [10–15].
In contrast to animals, which only maintain CG methylation, in most plants 5-meC occurs also in several sequence contexts (CG, CHG and CHH, where H is any of the bases A, T or C) and is catalyzed by different methyltransferases acting on different DNA methylation pathways. In A. thaliana, CG methylation is maintained by MET1, CHG methylation by CMT3 and CHH by CMT2 and the RNA-induced DNA methylation pathway. CG methylation occurs in euchromatin and heterochromatin whereas CHG and CHH methylation decorate repeats and transposons [16]. The cross-functioning and redundant DNA methylation pathways form a nuclear/DNA protection system that aids in identifying invading transposons and permanently shutting off their expression (see review by Kim et al. [17]).
Lister and Ecker [18] argued that 5-meC should be used as a dynamic fifth letter of the genomic code because of the important implications of methylation. It has become tractable to analyze genome-wide DNA methylation states in populations or across different plant species because of advances in next-generation sequencing (NGS) technologies. Much effort has been undertaken to determine the landscape of DNA methylation changes especially in A. thaliana and other land plants such as rice and tomato, which have had reference genomes available for several years [19, 20]. DNA methylation patterns vary widely among animals; Drosophila completely lacks CG methylation while the human genome is highly methylated (~75% of the cytosines). In A. thaliana, ~24% of the CGs, ~ 6.7% of the CHGs and ~1.7% of the CHHs are methylated [21, 22].
Plants have varying levels of repeat content, which might be the result of bursts of single-repeat retroelements, which can amplify rapidly using a reverse transcription step to make multiple copies, or DNA transposons, which use a copy-and-paste strategy [23, 24] and thus can amplify during DNA replication. While the repeat content is only ~20% in Arabidopsis, in cereals such as barley and wheat the repeat content can be up to 90%. Together with the presence of three subgenomes in hexaploid wheat, these repeats require tightly regulated epigenetic mechanisms [25]. Genes have evolved different mechanisms for tolerating transposable elements (TEs) in their vicinity [26, 27]. Hirsch and Springer [28] provide a review of the interactions between TEs and gene expression in plants. They discuss three mechanisms by which transposons influence gene expression, namely (i) the prevailing evidence that TE insertions within introns or untranslated regions of genes are often tolerated and have minimal impact on gene expression levels or splicing. Conversely, TE insertions within genes lead to aberrant or novel transcripts; (ii) TEs act as novel alternative promoters—with the potential to result in different expression patterns; and (iii) TE insertions near genes can influence gene regulation. In Arabidopsis, two genes (IBM1 and IBM2) have been identified that prevent spreading of CHG and CHH methylation from transposons into gene bodies or promoters.
Interestingly, DNA methylation levels can also affect how plants respond to stress. Arabidopsis mutants with reduced global DNA methylation show increased expression of defense-related genes and enhanced resistance to pathogens [29]. Polymorphisms of CMT2 correlate with DNA methylation variation along a longitudinal temperature gradient in natural populations [30], and cmt2 plants are more heat tolerant [31]. Isogenic lines with different DNA methylation states show differences in their ability to compete in synthetic plant communities [32]. Similar influences on stress tolerance have also been observed in monocots, and wheat with experimentally reduced DNA methylation shows resilience to salt and oxidative stress. The dynamics of the methylation state of genomic elements are tissue-specific (for instance, in A. thaliana seedlings [33–35]) and differ between juvenile and mature plants (e.g. in a study of Acacia mangium [36]). Reduced DNA methylation also results in abnormal plant development in A. thaliana [37]; hence, an optimally regulated level of methylation is vital for normal plant growth and development.
Plant pathogen invasion can also influence methylation levels in different ways. For instance, genome-wide hypomethylation and hypermethylation influence resistance-related genes [38] and alter gene expression profiles, resulting in plant adaptation to stress. Wang et al. [39] showed that drought-induced alterations to DNA methylation in rice influence an epigenetic mechanism that regulates gene expression. As a major modification of the eukaryotic genome, DNA methylation significantly influences gene expression. Methylation of genomic features can lead to different gene regulatory effects. For instance, alteration of a gene’s expression potential is a result of DNA methylation affecting the interaction between transcription factors and DNA with chromatin proteins [40]. Additionally, methylation of the promoter region results in repression of gene expression, and gene body methylation leads to the opposite effect [41, 42]. Studies have shown that gene body-methylated genes are constitutively expressed in a wide range of conditions and tissues [6].
Chemistry of bisulfite conversion and sequencing
Bisulfite sequencing is generally done in three main steps, namely (i) denaturing, (ii) bisulfite treatment and (iii) polymerase chain reaction (PCR) amplification. In bisulfite conversion, DNA is denatured in a process that separates the forward and reverse strands. This is followed by treatment with sodium bisulfite, which converts unmethylated cytosine into uracil—which is then converted to thymine during PCR [43]. Quantification of the abundance of each cytosine can be achieved via Sanger sequencing [44] or NGS technologies [45]. The DNA strands cease to be complementary after bisulfite conversion. Treatment of genomic DNA with sodium bisulfite [46] enables us to distinguish between highly similar (and yet different) methylated cytosine, which has the same base-pairing features as unmethylated cytosine. Mapping read sequences to a reference genome enables the determination of positions with matching and mismatching bases. This process enables identification of methylated and unmethylated bases.
Bisulfite sequencing can be accomplished with different sequencing kits depending on whether whole-genome bisulfite sequencing (WGBS) [18] or reduced-representation bisulfite sequencing (RRBS) [47, 48] is performed. Currently, WGBS remains the most informative method for generating DNA methylation data. It provides a huge wealth of data and requires no prior targeting. Unlike WGBS, which is expensive, RRBS can be performed more economically because it is restricted to CpG-enriched regions that make up a smaller portion of the genome. The restriction enzyme Msp1 cleaves at 5’-C*CGG-3’ targets (base preceding * is methylated), thereby, mainly CpG-rich regions are targeted—which is advantageous for large genomes.
Typical workflow for processing bisulfite sequencing data
Before reads are mapped to a reference genome, the sequencing quality of reads can be checked with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) or NGS QC Toolkit [49], followed by removing low-quality bases and adapters with, among others, Trim Galore (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore), cutadapt [50] or Trimmomatic [51]. However, some WGBS data processing tools integrate various analytic steps—enabling data preprocessing, read alignment, a more robust statistical analysis that output statistics such as read coverage, the percentage of uniquely aligned reads and statistics on the three methylation contexts (CpG/CHG/CHH). One such tool is gemBS [52], which is a recently published pipeline for processing and analysis of WGBS data. The pipeline integrates data preprocessing and analysis steps from adaptor trimming through downstream statistical analysis of mapping results. gemBS uses the high-performance read aligner GEM3 [53] as a dependency and BScall (embedded in samtools, bcftools; http://samtools.sourceforge.net), which is a variant caller for bisulfite sequencing data. Both GEM3 and BScall support single and paired-end reads. Further reading on the generic workflow of analyzing WGBS is found in the work of Liang et al. [54] and Wrecyzcka et al. [55].
Non-bisulfite-based methods and related bioinformatics tools
While bisulfite sequencing methods represent the most popular approaches for analyzing epigenomic data, there are other approaches within the field of DNA modification-based methods. These approaches include methylated DNA immunoprecipitation (MeDIP)-seq and MethylCap-seq (a robust procedure for genome-wide profiling of DNA methylation) in MeDIP analyses [56] where the genomic DNA is randomly sheared, sonicated and immunoprecipitated with an antibody recognizing 5-methylcytidine. Precipitated DNA can either be sequenced or hybridized to microarrays. MethylCap-seq uses the methyl-CpG-binding domain of MeCP2 [57] while Oxidative bisulfite sequencing (oxBS) [58] is used to specifically detect 5-methylcysteine and 5-hydroxymethylcytosine (5hmC) that can be also done with ‘Tet’-assisted bisulfite sequencing [59]. CAB and fCAB are used for the recognition of 5caC [60]. Notably, the presence/absence of 5hmC in plants remains contentious. Some scholars claim that 5hmC is present in plants [61, 62] while others claim it’s absent [63]. A comprehensive overview of the various tools is given at https://omictools.com/medip-seq-category.
Tools for analyzing epigenomics datasets
Bismark [64] and BSMap [65], as one of the 1st published tools for quantifying epigenomic datasets, had to address the challenge of attaining high-read mapping efficiency to enable a sensitive sequence search. Bowtie [66], Merman [67], SNAP (http://snap.cs.berkeley.edu) and Bowtie2 [68] have been used as dependencies in epigenomics tools, for instance, BS-Seeker [69], BS-Seeker2 [70], BS-Seeker3 [71], BRAT-nova [72], WALT [73] and Bismark, which are currently among the most commonly applied tools for mapping bisulfite methylation data. We outlined the most common tools for mapping bisulfite sequencing data along with tools that allow for the detection and analysis of differentially methylated regions (DMRs). The program parameters as well as input and output data formats are specified in Table S1. This table provides an overview of the main tools for mapping and analysis of epigenomic data—particularly for bisulfite sequencing data. Additionally, we also categorized the tools into three major classes, namely (i) mapping, (ii) statistical analysis and (iii) complete pipelines (Table S1). The defining features for each tool, such as their ability to handle single or double-stranded sequence data as well as their ability to process data and perform downstream statistical analysis, are also provided. Reviews by Adusulalli et al. [74], Shafi et al. [75] and Wrecyzcka et al. [55] complement our overview Table S1. The most frequently applied computational epigenetics methods were applied and tested using DNA methylation data, particularly with data acquired from bisulfite sequencing experiments. Therefore, there are many statistical procedures available for analyzing methylome data—categorized into the parametric and non-parametric approach. Both approaches are widely used in the literature [76]. For instance, MethylMix [77] is an excellent example of a parametric approach that uses Bayesian mixture models to identify DNA methylation states of genes as either hypo- or hypermethylated. The method entails fitting a distribution function onto the frequencies of DNA methylation counts. The advantage of using non-parametric models is that no prior knowledge of the data distribution is required. However, when such knowledge is available, then parametric models are the preferred choice for modeling such data. MethylMix quantifies the effect of DNA methylation on genes, which is interesting for integrative studies that aim at establishing the association between the methylation states of the individual genes and their expression profiles. Investigating such associations unravels any hidden variations within and between samples (or tissues) as illustrated in [78–80]. Lea et al. [81] discussed the applications of mixed models on DNA methylation in plant epigenetics. They specifically focused on the binomial mixed model with the sampling-based algorithm (MACAU, mixed model association for count data via data augmentation) for the approximation of parameters and computation of P-values.Other modeling frameworks are based on algorithms that integrate various analytical steps resulting in the detection of DMRs across the entire genome, for instance, (i) the weighted optimization algorithm proposed in [82] (which is an extension of MethylKit [83]) and (ii) ChAMP.DMR [84], which applies the Bumphunter [85] or ProbeLasso Algorithm [86]. An example of a non-parametric model is the Bayesian approach based on the Dirichlet process beta mixture model—which is used for clustering methylation profiles [76]. The model considers the DNA methylation expressions consisting of an infinite number of beta mixturex distributions [87, 88].
DNA methylation: plant physiology and pathophysiology
Investigating the dynamics of DNA methylation in plant growth and development requires the analysis of samples from different plant tissues (e.g. [34]). To our knowledge, no existing software has been developed specifically for the analysis of plant physiology and pathophysiology. However, there are many studies analyzing bisulfite data using samples from different plant developmental stages (from seedlings to mature plants). For instance, Bismark—in leaf tissues from bread wheat seedlings [89], BSMap—for various datasets from different tissues in A. thaliana [90] and BS-Seeker2—for young Zea mays leaves [91]. With rapid advancements in the development of software/tools for analysis of epigenomes, we are optimistic such tools will soon be available to the public.
DMRs and their significance
Genomic regions (or bases) with different methylation profiles between samples are known as DMRs. This is also referred to as differentially methylated CpG sites since the CpG-methylated sites occur in much larger numbers compared to the non-CpG contexts (CHG and CHH) [92, 93]. Peak detection enables the identification of CpG islands—which are essential for differentiating methylation profiles between samples (typically between controls and test samples). CpG islands are not randomly distributed in the genome but are instead grouped close together [94]. Long stretches of non-dense CpG sites, known as CpG shores, can also be detected. Combining the methylation profiles of both CpG-islands and CpG-shores enables more efficient comparative analysis of DNA methylation profiles between samples.
Various statistical algorithms have been proposed for identifying DMRs—the most popular ones being methylKit [83], metilene [95], DMRcaller [96] and Bumphunter [85]. For elaborate discussions on the DMR detection methods and a discussion on choosing the right method for DMR detection, see [97, 98]. The tools are written and compiled in different programming languages (e.g. R, Python, Perl, Java, C and C++; Table S1). Essentially, such tools are used to identify DMRs from either targeted regions of the genome or from the whole genome. Critical considerations have to be made, e.g. the choice of experimental designs for experiments and statistical methods for data analysis [99]. DMRs are intricately linked to transcription and the abundance of CpG sites (CpG islands). A high concentration of CpG sites is often found within the promoter regions of genes—so it is essential to accurately identify such sites. Methylation of promoter regions influences the level of transcription—heavy methylation disrupts transcription, and de-methylation leads to transcription reactivation [100–102].
Peak identification and normalization are crucial initial steps in analyzing DNA methylation data and visualization and can be useful for comparing datasets and judging the performance and agreement between tools. Post-processing and visualization of (differentially) methylated sites enable high-resolution exploration and comparison of regions in the genome for variations in methylation profiles. Therefore, tools like BiQ [103] and BSeQC [104] have aided quality control and visualization of methylation data, thereby enabling researchers to explore data attributes and perform data quality control before analysis. There are many methods for clustering methylation marks such as the dynamic genome warping [105] approach that uses hierarchical clustering and the combination of different epigenomics analytic platforms and data integrative modules. Dynamic genome warping has been demonstrated to be a reliable way to get more meaningful results from datasets (for instance, [106]). To utilize this method, Liang et al. [54] developed a webserver to analyze WGBS data and their platform includes major steps for detection and mapping of the conversion rate, detection of DMRs and their association with gene expression. Wreczycka et al. [55] discussed data requirements and computational attributes for specific software and assess bisulfite sequencing data analysis methods, alignment and data processing, detection of differential methylation and assess strategies for handling large epigenetic datasets. In contrast, our work highlights existing asymmetries between mapping tools and contrasts their computational capabilities.
Another important aspect in plant epigenetics is how hypomethylation and hypermethylation affects gene expression. The concept of hypomethylation and hypermethylation is not limited to plants as they have also been extensively studied in cancer progression in humans [107], coronary heart disease [108] and eukaryotes in general [109]. The division of DMRs into hypo- and hypermethylated enables investigations into the influence of both types of methylation on gene expression. Many computational tools have integrated modules that enable the extraction and quantification of the extent of hypo- and hypermethylation in genes. One such tool is MethylMix, which requires that changes in a gene’s methylation state must also agree with its expression profile. Additionally, it requires a treatment and control sample (for agricultural studies) or healthy and disease conditions (for clinical studies).
Downstream analyses of bisulfite methylome data
After data processing and calling of methylation sites, down-stream analysis can be performed—including the functional annotation of DMRs and analysis of the associated pathways influenced by the targeted genes. Such analysis enables the assignment of functions and gene annotation as seen in the overviews of Bioinformatics omicX tools (https://omictools.com/epigenomics-category). Examples of tools for performing down-stream analysis are given in Table 1.
Table 1. Examples of some downstream analysis software.
| Tool | Citation and descriptions |
|---|---|
| ADMIRE: Analysis and visualization of differential methylation in genomic regions using the Infinium HumanMethylation450 Assay | Preussner et al. [109]; online and offline. Adds experimental settings, quality control, automatic filtering, normalization, multiple testing and differential analyses genome browser tracks, table outputs and summary files. |
| BATMAN: Bayesian automated metabolite analyser for Nuclear magnetic resonance (NMR) spectra | Hao et al. [110]; uses Markov chain Monte Carlo algorithm for sampling. Bayesian-based approach. |
| KEGG: Gene Ontology Pathways | It is a database for mining and analysis of high-level functions. KEGG enables analysis and data mining on different biological scales (e.g. cellular and molecular-level information, whole organism, at ecosystem level, etc—using data from high-throughput experiments; see https://www.genome.jp/kegg). |
| IPA: Ingenuity Pathway Analysis | Krämer et al. [111]; platform enables exploration and visualization of complex omics data (e.g. microarrays including miRNA, metabolomics, proteomics, Ribonucleic acid sequencing (RNA-Seq), small RNA-Seq and single-nucleotide polymorphism (SNP) and small-scale experiments); see https://www.qiagenbioinformatics.com. |
| DAVID: Database for Annotation, Visualization and Integrated Discovery | Huang et al. [112]; DAVID enables pathway mining and gene function classification. Input is gene list from high-throughput genomic experiments; see https://david.ncifcrf.gov. |
Technical challenges: conversion rate, repetitive regions and DMRs
The main challenges in the analysis of DNA methylation data include incomplete methylation patterns and overdispersion of read mappings [110–112]. Here, overdispersion means the presence of variability in the reads compared to the expected read distributions based on a given model structure. When epigenomics marks coincide with repetitive regions in the genome, mapping tools need to keep reads that map to multiple genomic locations—making these tools slower and computationally memory-intensive. This problem can be partly circumvented through parallel computing using multiple threads, especially for larger repetitive plant genomes.
Conversion rates
As a method for studying DNA methylation, bisulfite conversion involves the conversion of cytosine to uracil (while 5-methylcytosine, 5-mC remains unchanged). Bisulfite sequence conversion rates vary for different datasets. It is essential for conversion rates to be determined accurately to ensure the reliability of downstream data analysis. Reliable results can be obtained from datasets with bisulfite conversion rates higher than ~0.999 (see, e.g. [113]—demonstrated using their tool MethQA). However, they urge caution for datasets with lower conversion rates. Modern commercially available bisulfite sequence conversion kits generally indicate conversion efficiencies of 90–100% [114]. An elaborate discussion on methods for estimating conversion rate from bisulfite DNA methylation data is provided in [115, 116].
Description of experiment: benchmarking selected tools
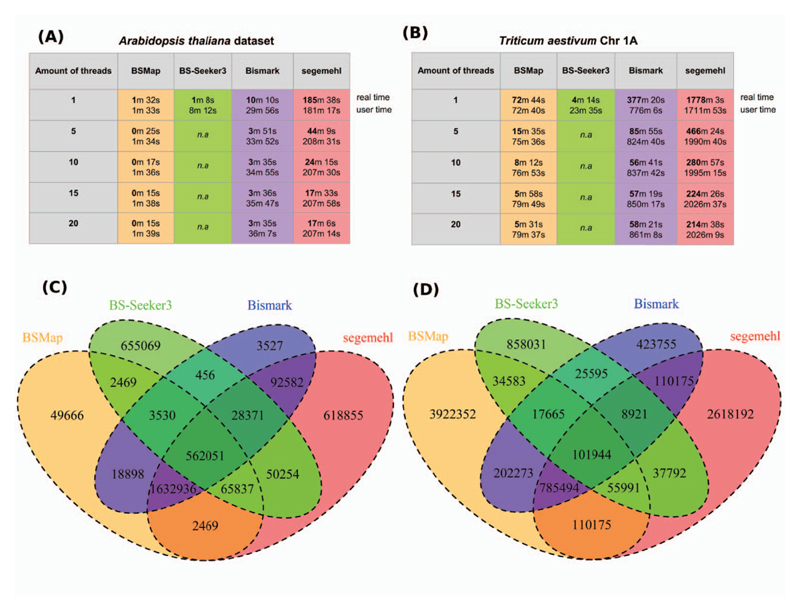
We aimed to determine how the well-established computational epigenomics methods perform on a small genome such as A. thaliana with ~130 Mbp (TAIR10) compared to a genome with a high repeat content and much larger genome size such as bread wheat—taking chromosome 1A (Chr1A) for demonstration purpose [117]. We used bisulfite sequencing data from two studies (with accession numbers SRR429549 [118, 119] for A. thaliana and ERR1141918 [89] for Triticum aestivum; data from NCBI) and applied four methods: BSMap [65], Bismark [64], BS-Seeker3 and segemehl [120]. Our analysis focused on the speed and agreement of common methylated sites between the tools. BS-Seeker3 was the fastest, followed by BSMap, while Bismark and segemehl were the slowest irrespective of genome size—especially for multiple threads (Figure 1A and B). When using a single thread, segemehl (keeping reads that mapped a maximum of three times) performed slowest compared with the other methods. Overall, the computation time required for the T. aestivum (Chr1A) dataset is significantly longer than those from A. thaliana (Figure 1A and B). When comparing the reported sites, we found that, for A. thaliana, 562 051 sites are shared among all four tools. While most sites were overlapping between BSMap, BS-Seeker3 and Bismark, likely because they use the same mapping software, segemehl reported only ~10% of these sites. However, for T. aestivum, ~101 944 sites were reported with most of them being reported in segemehl (Figure 1C and D). The existence of such asymmetries requires more attention and is certainly worth taking into consideration when using the different computational tools. Other studies on comparisons of the performance of epigenetics analysis tools, specifically focusing on mapping short reads for bisulfite sequencing data, can be found in the work of Tran et al. [121]. Several studies have also compared runtime and memory consumption of different epigenomics tools, such as Tran et al. [121] who compared the five bisulfite short read-mapping tools BSMap, Bismark, BS-Seeker, Bisulfite Sequencing Scorer (BiSS) and BRAT-BW and Bismark performed best on real data, followed by BiSS, BSMap and BRAT-BW and BS-Seeker. Recently, Huang et al. [71] proposed BS-Seeker3—a fast mapping tool for bisulfite data and compared it performance for runtime and sensitivity to sister tools like Bismark, BRAT-nova and BSMap. Additional to being accurate and versatile, Huang et al. concluded that BS-Seeker3 is an ultra-fast pipeline to process bisulfite-converted reads. The tool also aids visualization of methylation data, hence justifying its comparability to the other three tools (Bismark, BRAT-nova and BSMap).
Figure 1.
Selection of epigenomics tools. (A and B) Results of the calculation user times for four common tools, Bismark, BSMap, BS-Seeker3 and segemehl. We used data for A. thaliana and chromosome 1A in bread wheat (T. aestivum). n.a, values not available. (C and D) Overlap of detected sites in the two reference genomes for the four mapping tools.
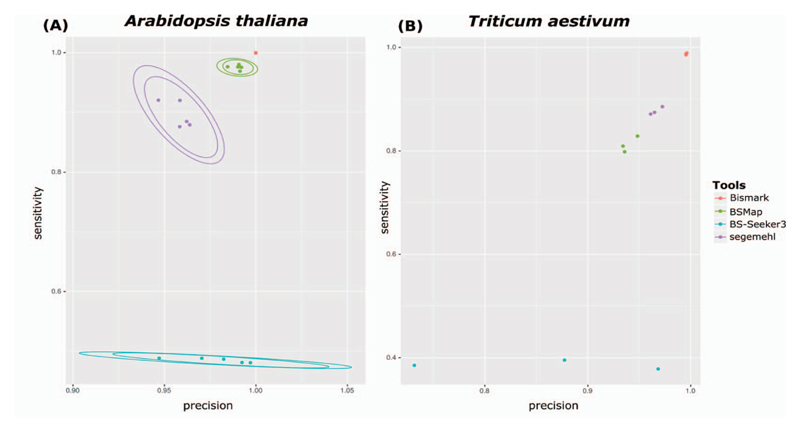
We simulated reads from A. thaliana and bread wheat using the tool by Sherman (https://www.bioinformatics.babraham.ac.uk/projects/sherman) to test the performances of the four tools by comparing the precision and sensitivity along all chromosomes (Figure 2). The sensitivity, also sometimes referred to as recall, is defined as TP/(TP + FN). The precision is defined as TP/(TP + FP), where TP—true positive, FN—false negative and FP—false positive. We observed best performances for the Bismark, followed by BSMap and segemehl, while BS-Seeker3 seemed to have a lower sensitivity in A. thaliana compared to the other tools. For bread wheat a similar order to performances of tools was observed when reads where simulated for each subgenomes of chromosome 1 with the three genome copies. All scripts were provided in GitHub (https://github.com/jomony/EPItools/blob/master/README.md).
Figure 2.
Precision and sensitivity analysis. Precision and sensitivity analysis for the A. thaliana data based on read mapping of simulated reads using the tool by Sherman (https://www.bioinformatics.babraham.ac.uk/projects/sherman)—with the parameters (CG = 24, CH = 8, e = 0.5). (A) There is a large difference in the sensitivity of the four tools. BS-Seeker3 was the least sensitive (sensitivity averaging ~48%)—Bismark was the most sensitive (sensitivity, ~99.9%). The sensitivity values for BSMap and segemehl averaged ~97% and 90%, respectively. (B) For bread wheat (T. aestiuum), BSMap appears to be marginally less precise and less sensitive than segemehl. There is consistency in the precision and sensitivity values for the subgenomes A, B and D in chromosome 1 of T. aestivum. Overall, the results from both (A) and (B) are in agreement. Notably, BS-Seeker3 has a wide range of precision compared to the other three tools. Each data point represents the precision-sensitivity value based on a simulation run for an individual tool. The precision and sensitivity values for Bismark, BSMap, BS-Seeker3 and segemehl averaged ~(99%, 99%), (94%, 82%), (86%, 38%) and (97%, 87%), respectively. Five simulation runs were performed for each tool—one for each of the A. thaliana chromosomes. The elliptical rings around each set of data points represent the confidence bounds.
Feature comparison between the tools and related literature benchmarking
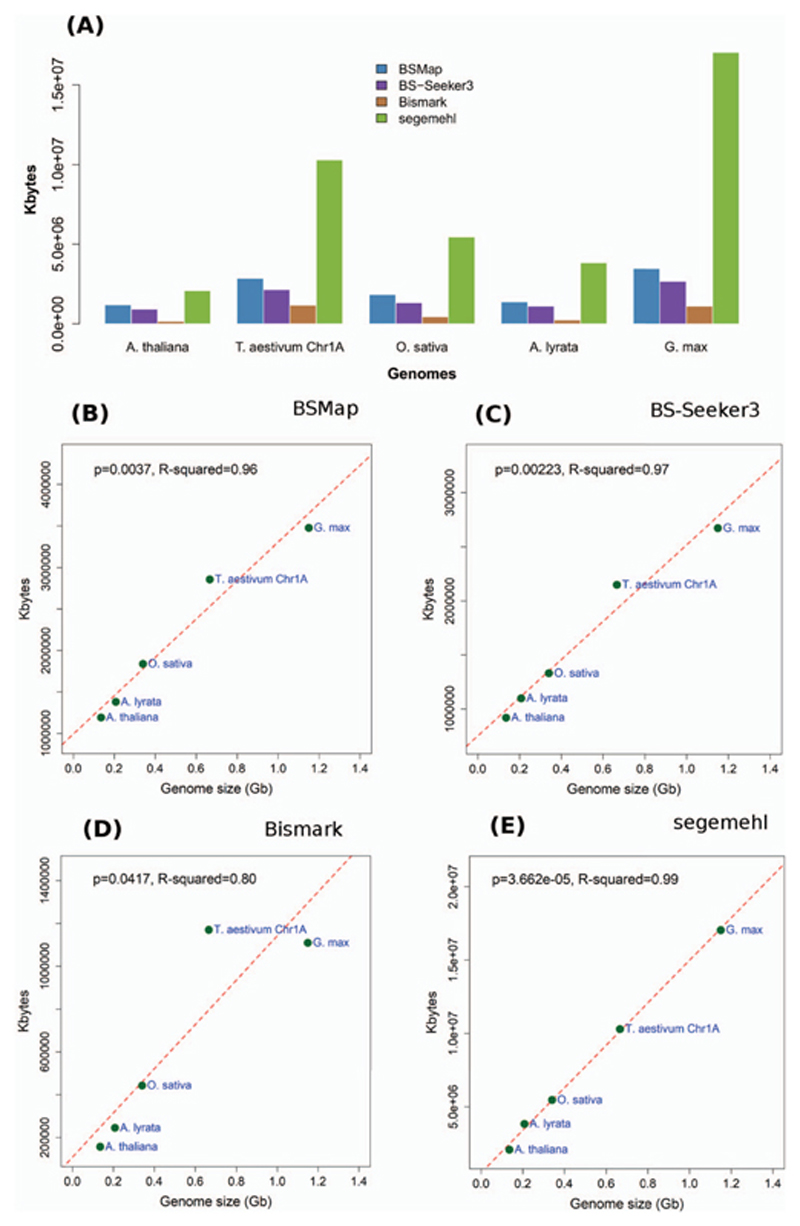
To further benchmark the performance of the tools, we used bisulfite sequencing data from five plant genomes. These genomes consist of the dicots: A. thaliana (genome size, ~0.13 Gb; SRR4295494), Arabidopsis lyrata (~0.21 Gb; SRR3880297) and Glycine max (~1.2 Gb, SRR5079790) and also the monocots: T. aestivum (chromosome 1A; size, ~0.67 Gb; ERR1141918) and Oryza sativa (~0.43Gb; SRR7265433). Figure 3A shows the results of a comparative analysis of the memory footprint analysis of the performance of the four tools benchmarked using data from five genomes. These results come from mapping the bisulfite reads data to their respective reference genomes. Association analysis was performed for each of the four tools as seen in the linear regression model fits (Figure 3B–E). The results show that the genome sizes for each of the five genomes are significantly correlated to the memory footprint analysis (P < 0.05).
Figure 3.
Memory footprint analysis for the four tools—benchmarked on five genomes. (A) Barplots showing variation in attained memory footprint between the tools benchmarked on different genomes. (B−E) Correlation analysis of genome size and memory footprint analysis. A benchmark of the four tools, (B) BSMap, (C) BS-Seeker3, (D) Bismark and (E) segemehl. The genome sizes are all significantly correlated to the memory footprint analysis (P < 0.05). Dotted line, fitted regression line; Dots, data points.
The key attributes and parameters for the four tools are summarized in Table S2. This table presents a summary of the supported features in the four tools (BSMap, BS-Seeker3, Bismark and segemehl). Such features are essential for deciding on which tool to use for mapping reads and data analysis. Examples of such features can also be found in the work of Guo et al. [70] and Tran et al. [121]. Lee et al. [122] evaluated the mapping accuracy and mapping rates for Bismark, BSMap and BS-Seeker2 as a function of the error rates. Using WGBS data, they assessed the influence of the error rates on the mapping rates and mapping accuracy and observed that at low error rates (<4%), BSMap had a higher mapping rate than Bismark and BS-Seeker2. On the contrary, BSMap had a lower mapping accuracy than Bismark and BS-Seeker2. They also showed that mapping accuracy is independent of the methylation level.
A discussion on benchmarking approaches with a focus on short sequence mapping tools is found in the work of Hatem et al. [123]. They assess the performance of various aligners for the read mapping tools and benchmark them using criteria such as mapping percentage, running time and memory footprint. Variations in parameters such as seed length, base quality and single- or paired-end reads on the mapping quality are also evaluated. Benchmarking of tools by comparing the performance of each tool based on multiple attributes can be achieved in various ways, for instance, by assessing (i) the effect of the read length and sequencing error, (ii) the effect of data processing and (iii) the effect of varying parameters in the tools. These are some of the approaches discussed by Tran et al. [121]. They compared the performance of epigenomic mapping tools such as BSMap, Bismark, BS-Seeker, BRAT-BW [124] and the BiSS [125]. Tran et al. primarily benchmarked the performance of the tools basing on mapping efficiency (as the percentage of reads that map uniquely to the genome) and the central processing unit (CPU) time.
Outlook
In the near future, there is a need for more comparative analyses to explore the epigenomes of diverse plants in different development stages together with various stress factors. This would enable the discovery of exclusive and common epigenetic regulatory mechanisms. Uncovering and exploiting such mechanisms could potentially promote adaptation to changing environmental conditions. Moreover, a large number of methylomes are required to study the effect of the environment and stress conditions on the epigenomic state of a single plant [126, 127]. Resources like the 1001 Epigenomes Project (https://1001genomes.org) in A. thaliana are exciting datasets to aid in our understanding of the role of the epigenome. However, it remains unclear whether the observations in these studies are directly applicable to crops.
Computational tools are instrumental for bridging the gap between mapping of sequenced reads, the accurate prediction of methylated sites and their statistical analysis However, this effort is hampered by variations in the size of epigenomic marks and the complexity associated with normalizing peaks. The need to increase crop yield on the same amount, and in some cases dwindling, of arable land is another important aspect that requires advancements in epigenomics studies. Several studies have shown that during seed and grain development, the plant epigenome changes and leads to gene silencing. Therefore, a change in the epigenetic state of a plant would result in an increase in its likelihood of adapting from one geographical location to another or to different environmental conditions.
Lämke and Bäur [128] argued that such modifications have the potential to provide a mechanistic basis for stress memory in plants. This enables plants to respond more efficiently to recurring stress from the environment, for instance, drought and salinity stress [129], a topic that was reviewed by Golldack et al. [130] (and more recently by Yang and Guo [131] and Abhinandan et al. [132]). This might enable plants to prepare their offspring for future attacks from stressors and to improve their adaptation to specific stress factors [130]. Plant adaptation to stress might enable us to explore new ways to improve yield, for instance, by shortening or prolonging the time for grain development, by finding ways to regulate the expression of the three homeologs in wheat or by interfering with fruit ripening (as seen in tomatoes [133–135] and other fruits like peach, apples and strawberries [136]). A more intriguing discussion on the epigenetic mechanisms of plant stress response and adaptation to different environmental conditions was reviewed in [137–139].
In this review, we have discussed the use of bioinformatics tools to study DNA methylation data in plants. Notably, several studies in humans and mouse were successfully performed using popular tools like BSMap, BS-Seeker/BS-Seeker2/BS-Seeker3, Bismark in mouse and segemehl in human cancer cell lines. For the analysis of bisulfite sequence data, most of the fundamentals of the chemical background and methylation principles are the same; however, the major difference between the use of such tools in plants and animals (specifically, in humans and mouse) is the genome structure organization and the presence of predominantly more CHG/CHH methylation contexts in plants. The most predominant context of DNA methylation in mammals is the symmetric CG—estimated to be at ~70−80% of CG dinucleotides genome-wide [140]. The mechanisms of regulation and function of DNA methylation vary in animals and plants [141, 142]. These variations in regulation and function mechanism, coupled with genome structure differences and complexity levels, is a motivating factor for integrating small subtle differences in mapping and analysis tools for epigenome data. Another important difference of plants and animals is how they are able to demethylate their genome. So far, enzymes removing directly the methyl group from cytosines have not been identified in plants, but they are important components of mammalian DNA methylation homeostasis. Plants use either passive mechanisms (not maintaining methylation during DNA replication) or base excision and subsequent repair for direct removal of methylated cytosines. Unlike with the human genome, the CHG/CHH contexts that are more abundant in plants [143] need to be integrated into the mapping and analysis of methylome data. Many plants have large and repetitive genomes compared to that of humans. Such large genomes are a limiting factor in the analysis since they require a lot of computational resources. The sequence mapping to references and statistical computational time for large genomes such that of bread wheat (~17 Gb) and barley (~5.3 Gb) is likely to scale linearly.
Concluding remarks
In the last decade, there has been tremendous progress in the development of tools for analyzing epigenomic data; however, numerous challenges remain. For instance, the visualization capacity of many tools remains either inadequate or lacks essential modules for handling and displaying statistical outcomes from the resulting analysis. Additionally, the ability of these tools to scale-up and to handle large genomes remains an issue for further exploration. Technically, most computational tools for analyzing epigenomic data perform well for datasets from organisms with a genome size that is smaller than the human genome (~3 Gb). For much larger and complex genomes, more computational resources are required, and the genome structure (whether diploid, hexaploidy or tetraploid) and repetitive nature of the genome have to be taken into consideration during mapping to a reference genome. This is demonstrated in our example where we compared the mapping efficiency for Arabidopsis and a wheat chromosome; however, the complexity in genome structure, the presence of TEs and the lack of consistent gene annotations for some plants remain a major obstacle to advancing epigenetic research.
In the next decade, there is likely to be a steady improvement in sequencing methods and performance of already existing computational algorithms. Recently, it was shown that even well-established sequencing methods might be prone to errors, leading to misleading results, e.g. DNA immunoprecipitation sequencing [144]. Discovering and amending such errors can lead to new findings from the previous studies and limit these errors’ damage to future studies. This will aid further epigenetic research not only in plants but also in life sciences in general. Additionally, a few tools have the capability to effectively get more information out of low-coverage data. Developing new tools or improving on existing ones to attain optimal results using low coverage data and fewer replicates would save experiment and sequencing costs. A high sequence coverage allows for good data quality and enables robust statistical analysis [145]. Achieving high sequence coverage can be quite expensive and the minimum desired coverage can depend on the research objectives at hand. Typically, a coverage value of 5–10 × is sufficient for many comparative studies and for achieving reliable methylation calls [145]. However, studies have demonstrated that coverage values as low as 2× is sufficient [146]. Accurate identification of DMRs in large samples, especially between multiple conditions, remains a challenge—despite tremendous progress already made in this area.
Supplementary Material
Key Points.
We introduce the concepts of epigenetics in plants and discuss commonly used tools—with a focus on their capabilities.
Integration of bioinformatics tools needed to understand epigenomics datasets in crops.
The presence of repetitive elements in the genome influences the prediction of methylated sites.
We list the runtime and computational requirement for a small and large complex genome and demonstrate their overlaps in four most applied tools.
Different tools have different levels of asymmetry with regards to their mapping and methylation call statistics.
Acknowledgements
We would like to acknowledge the de.NBI infrastructure for the possibility to run the various methods. We extend our appreciation to Manuel Spannagl and Klaus F.X. Mayer (of PGSB, Helmholtz Zentrum München) for their suggestions on stream-lining the manuscript. J.O was supported by the “INTREPID project”. We thank Ortrun Mittelsten Scheid, and J. Matthew Watson (both GMI, Vienna) for helpful comments on the manuscript. We gratefully acknowledge financial support from the Plant Fellows program (EU FP7 to R.G.) and the Austrian Science Fund (FWF I3687-B25 to R.G.) as part of the ERA-CAPS “AUREATE” consortium project.
Footnotes
Author Contributions
J.O. and T.N. initiated the study, initiated the analysis frame-work, performed the data analysis and simulations and drafted the manuscript. R.G. streamlined the manuscript content and write-up.
Contributor Information
Jimmy Omony, postdoc (bioinformatician) at the Plant Genome and Systems Biology, Helmholtz Center Munich, Germany. His research interests include plant genomics, epigenetics, machine learning and biostatistics. He undertook the first postdoc at the University of Groningen (RuG). He holds a PhD in computational systems biology (Wageningen University).
Thomas Nussbaumer, postdoc (bioinformatician) at the Institute of Network Biology and also in the Institute of Environmental Medicine. His research interests include epigenomics, plant genomics, protein-protein interaction analysis and microbiomics. He undertook his first postdoc at the University of Vienna and is currently a postdoc Fellowship Program holder at the Helmholtz Center Munich.
Ruben Gutzat, postdoc at the Gregor Mendel Institute of Molecular Plant Biology, Austrian Academy of Sciences, Vienna (Austria). His research interests are in plant epigenetics and developmental biology.
References
- 1.Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38(5):285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takuno S, Ran JH, Gaut BS. Evolutionary patterns of genic DNA methylation vary across land plants. Nat Plants. 2016;2:15222. doi: 10.1038/nplants.2015.222. [DOI] [PubMed] [Google Scholar]
- 3.Bewick AJ, Ji L, Niederhuth CE, et al. On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci U S A. 2016;113(32):9111–6. doi: 10.1073/pnas.1604666113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bewick AJ, Schmitz RJ. Gene body DNA methylation in plants. Curr Opin Plant Biol. 2017;36:103–10. doi: 10.1016/j.pbi.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Wang X, Lee T-H, et al. Gene body methylation shows distinct patterns associated with different gene origins and duplication modes and has a heterogeneous relationship with gene expression in Oryza sativa (rice) New Phytol. 2013;198(1):274–83. doi: 10.1111/nph.12137. [DOI] [PubMed] [Google Scholar]
- 6.Bewick AJ, Vogel KJ, Moore A, et al. Evolution of DNA methylation across insects. Mol Biol Euol. 2017;34(3):654–65. doi: 10.1093/molbev/msw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauss K, Wardenaar R, Oka R, et al. Parental DNA methylation states are associated with heterosis in epigenetic hybrids. Plant Physiol. 2018;176(2):1627–45. doi: 10.1104/pp.17.01054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kooke R, Keurentjes JJ. Epigenetic variation contributes to environmental adaptation of Arabidopsis thaliana. Plant Signal Behau. 2015;10(9):e1057368. doi: 10.1080/15592324.2015.1057368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortijo S, Wardenaar R, Colomé-Tatché M, et al. Mapping the epigenetic basis of complex traits. Science. 2014;343(6175):1145–8. doi: 10.1126/science.1248127. [DOI] [PubMed] [Google Scholar]
- 10.Kakutani T. Epi-alleles in plants: inheritance of epigenetic information over generations. Plant Cell Physiol. 2002;43(10):1106–11. doi: 10.1093/pcp/pcf131. [DOI] [PubMed] [Google Scholar]
- 11.Quadrana L, Colot V. Plant transgenerational epigenetics. Annu Rev Genet. 2016;50:467–91. doi: 10.1146/annurev-genet-120215-035254. [DOI] [PubMed] [Google Scholar]
- 12.Hauser MT, Aufsatz W, Jonak C, et al. Transgenerational epigenetic inheritance in plants. Biochim Biophys Acta. 2011;1809(8):459–68. doi: 10.1016/j.bbagrm.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossniklaus U, Kelly WG, Kelly B, et al. Transgenerational epigenetic inheritance: how important is it? Nat Rev Genet. 2013;14(3):228–35. doi: 10.1038/nrg3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martienssen RA, Colot V. DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science. 2001;293(5532):1070–4. doi: 10.1126/science.293.5532.1070. [DOI] [PubMed] [Google Scholar]
- 16.Deleris A, Stroud H, Bernatavichute Y, et al. Loss of the DNA methyltransferase MET1 induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 2012;8(11):e1003062. doi: 10.1371/journal.pgen.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YJ, Lee J, Han K. Transposable elements: no more ‘Junk DNA’. Genomics Inform. 2012;10(4):226–33. doi: 10.5808/GI.2012.10.4.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19(6):959–66. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomato Genome C. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–41. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goff SA, Ricke D, Lan TH, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296(5565):92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 21.Lanciano S, Mirouze M. DNA methylation in rice and relevance for breeding. Epigenomes. 2017;1(2):10. [Google Scholar]
- 22.Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wicker T, Sabot F, Hua-Van A, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 24.Tsukahara S, Kobayashi A, Kawabe A, et al. Bursts of retrotransposition reproduced in Arabidopsis. Nature. 2009;461:423. doi: 10.1038/nature08351. [DOI] [PubMed] [Google Scholar]
- 25.Eichten SR, Stuart T, Srivastava A, et al. DNA methylation profiles of diverse Brachypodium distachyon align with underlying genetic diversity. Genome Res. 2016;26(11):1520–31. doi: 10.1101/gr.205468.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubin MJ, Mittelsten Scheid O, Becker C. Transposons: a blessing curse. Curr Opin Plant Biol. 2018;42:23–9. doi: 10.1016/j.pbi.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Bourque G, Burns KH, Gehring M, et al. Ten things you should know about transposable elements. Genome Biol. 2018;19(1):199. doi: 10.1186/s13059-018-1577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch CD, Springer NM. Transposable element influences on gene expression in plants. Biochim Biophys Acta Gene Regul Mech. 2017;1860(1):157–65. doi: 10.1016/j.bbagrm.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Dowen RH, Pelizzola M, Schmitz RJ, et al. Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci U S A. 2012;109(32):E2183–91. doi: 10.1073/pnas.1209329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubin MJ, Zhang P, Meng D, et al. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. Elife. 2015;4:e05255. doi: 10.7554/eLife.05255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen X, De Jonge J, Forsberg SK, et al. Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet. 2014;10(12):e1004842. doi: 10.1371/journal.pgen.1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bossdorf O, Prati D, Auge H, et al. Reduced competitive ability in an invasive plant. Ecol Lett. 2004;7:346–53. [Google Scholar]
- 33.Bouyer D, Kramdi A, Kassam M, et al. DNA methylation dynamics during early plant life. Genome Biol. 2017;18(1):179. doi: 10.1186/s13059-017-1313-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartels A, Han Q, Nair P, et al. Dynamic DNA methylation in plant growth and development. Int J Mol Sci. 2018;19(7):E2144. doi: 10.3390/ijms19072144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang M, Kimatu JN, Xu K, et al. DNA cytosine methylation in plant development. J Genet Genomics. 2010;37(1):1–12. doi: 10.1016/S1673-8527(09)60020-5. [DOI] [PubMed] [Google Scholar]
- 36.Baurens FC, Nicolleau J, Legavre T, et al. Genomic DNA methylation of juvenile and mature Acacia mangium micropropagated in vitro with reference to leaf morphology as a phase change marker. Tree Physiol. 2004;24(4):401–7. doi: 10.1093/treephys/24.4.401. [DOI] [PubMed] [Google Scholar]
- 37.Finnegan EJ, Peacock WJ, Dennis ES. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA. 1996;93(16):8449–54. doi: 10.1073/pnas.93.16.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng H, Zhang J. Plant genomic DNA methylation in response to stresses: potential applications and challenges in plant breeding. Prog Nat Sci. 2009;19(9):1037–45. [Google Scholar]
- 39.Wang W, Qin Q, Sun F, et al. Genome-wide differences in DNA methylation changes in two contrasting rice genotypes in response to drought conditions. Front Plant Sci. 2016;7:1675. doi: 10.3389/fpls.2016.01675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55(3):451–8. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zilberman D, Gehring M, Tran RK, et al. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39(1):61. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Zhu J, Hu F, et al. Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics. 2012;13(1):300. doi: 10.1186/1471-2164-13-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marx V. Genetics: profiling DNA methylation and beyond. Nat Methods. 2016;13(2):119–22. doi: 10.1038/nmeth.3736. [DOI] [PubMed] [Google Scholar]
- 44.Eckhardt F, Lewin J, Cortese R, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38(12):1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yong WS, Hsu FM, Chen PY. Profiling genome-wide DNA methylation. Epigenetics Chromatin. 2016;9:26. doi: 10.1186/s13072-016-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark SJ, Harrison J, Paul CL, et al. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22(15):2990–7. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeddeloh JA, Greally JM, Rando OJ. Reduced-representation methylation mapping. Genome Biol. 2008;9(8):231. doi: 10.1186/gb-2008-9-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt M, Van Bel M, Woloszynska M, et al. Plant-RRBS, a bisulfite and next-generation sequencing-based methylome profiling method enriching for coverage of cytosine positions. BMC Plant Biol. 2017;17(1):115. doi: 10.1186/s12870-017-1070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7(2):e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Bioinform Action. 2012;17(1):10–2. [Google Scholar]
- 51.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merkel A, Fernandez-Callejo M, Casals E, et al. gemBS—high throughput processing for DNA methylation data from bisulfite sequencing. Bioinformatics. 2018;35(5):737–42. doi: 10.1093/bioinformatics/bty690. [DOI] [PubMed] [Google Scholar]
- 53.Marco-Sola S, Sammeth M, Guigo R, et al. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods. 2012;9(12):1185–8. doi: 10.1038/nmeth.2221. [DOI] [PubMed] [Google Scholar]
- 54.Liang F, Tang B, Wang Y, et al. WBSA: web service for bisulfite sequencing data analysis. PLoS One. 2014;9(1):e86707. doi: 10.1371/journal.pone.0086707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wreczycka K, Gosdschan A, Yusuf D, et al. Strategies for analyzing bisulfite sequencing data. J Biotechnol. 2017;261:105–15. doi: 10.1016/j.jbiotec.2017.08.007. [DOI] [PubMed] [Google Scholar]
- 56.Mohn F, Weber M, Schubeler D, et al. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 57.Brinkman AB, Simmer F, Ma K, et al. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52(3):232–6. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 58.Booth MJ, Ost TW, Beraldi D, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc. 2013;8(10):1841–51. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu M, Hon GC, Szulwach KE, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149(6):1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu X, Song CX, Szulwach K, et al. Chemical modification-assisted bisulfite sequencing (CAB-Seq) for 5-carboxylcytosine detection in DNA. J Am Chem Soc. 2013;135(25):9315–7. doi: 10.1021/ja4044856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang XL, Song SH, Wu YS, et al. Genome-wide mapping of 5-hydroxymethylcytosine in three rice cultivars reveals its preferential localization in transcriptionally silent transposable element genes. J Exp Bot. 2015;66(21):6651–63. doi: 10.1093/jxb/erv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shi DQ, Ali I, Tang J, et al. New insights into 5hmC DNA modification: generation, distribution and function. Front Genet. 2017;8:100. doi: 10.3389/fgene.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Erdmann RM, Souza AL, Clish CB, et al. 5-hydroxymethylcytosine is not present in appreciable quantities in Arabidopsis DNA. G3 (Bethesda) 2014;5(1):1–8. doi: 10.1534/g3.114.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics. 2011;27(11):1571–2. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hansen KD, Langmead B, Irizarry RA. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012;13(10):R83. doi: 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen PY, Cokus SJ, Pellegrini M. BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics. 2010;11:203. doi: 10.1186/1471-2105-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo W, Fiziev P, Yan W, et al. BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics. 2013;14:774. doi: 10.1186/1471-2164-14-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang KYY, Huang YJ, Chen PY. BS-Seeker3: ultrafast pipeline for bisulfite sequencing. BMC Bioinformatics. 2018;19(1):111. doi: 10.1186/s12859-018-2120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harris EY, Ounit R, Lonardi S. BRAT-nova: fast and accurate mapping of bisulfite-treated reads. Bioinformatics. 2016;32(17):2696–8. doi: 10.1093/bioinformatics/btw226. [DOI] [PubMed] [Google Scholar]
- 73.Chen H, Smith AD, Chen T. WALT: fast and accurate read mapping for bisulfite sequencing. Bioinformatics. 2016;32(22):3507–9. doi: 10.1093/bioinformatics/btw490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adusumalli S, Mohd Omar MF, Soong R, et al. Methodological aspects of whole-genome bisulfite sequencing analysis. Brief Bioinform. 2015;16(3):369–79. doi: 10.1093/bib/bbu016. [DOI] [PubMed] [Google Scholar]
- 75.Shafi A, Mitrea C, Nguyen T, et al. A survey of the approaches for identifying differential methylation using bisulfite sequencing data. Brief Bioinform. 2017;19(5):737–53. doi: 10.1093/bib/bbx013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang L, Meng J, Liu H, et al. A nonparametric Bayesian approach for clustering bisulfate-based DNA methylation profiles. BMC Genomics. 2012;13(6):S20. doi: 10.1186/1471-2164-13-S6-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cedoz PL, Prunello M, Brennan K, et al. MethylMix 2.0: an R package for identifying DNA methylation genes. Bioinformatics. 2018;34(17):3044–6. doi: 10.1093/bioinformatics/bty156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Widman N, Feng S, Jacobsen SE, et al. Epigenetic differences between shoots and roots in Arabidopsis reveals tissue-specific regulation. Epigenetics. 2014;9(2):236–42. doi: 10.4161/epi.26869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turco GM, Kajala K, Kunde-Ramamoorthy G, et al. DNA methylation and gene expression regulation associated with vascularization in Sorghum bicolor. New Phytol. 2017;214(3):1213–29. doi: 10.1111/nph.14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gardiner LJ, Joynson R, Omony J, et al. Hidden variation in polyploid wheat drives local adaptation. Genome Res. 2018;28(9):1319–32. doi: 10.1101/gr.233551.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lea AJ, Tung J, Zhou X. A flexible, efficient binomial mixed model for identifying differential DNA methylation in bisulfite sequencing data. PLoS Genet. 2015;11(11):e1005650. doi: 10.1371/journal.pgen.1005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li S, Garrett-Bakelman FE, Akalin A, et al. An optimized algorithm for detecting and annotating regional differential methylation. BMC Bioinformatics. 2013;14(5):S10. doi: 10.1186/1471-2105-14-S5-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Akalin A, Kormaksson M, Li S, et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tian Y, Morris TJ, Webster AP, et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics. 2017;33(24):3982–4. doi: 10.1093/bioinformatics/btx513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41(1):200–9. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Butcher LM, Beck S. Probe Lasso: a novel method to rope in differentially methylated regions with 450K DNA methylation data. Methods. 2015;72:21–8. doi: 10.1016/j.ymeth.2014.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58(301):236–44. [Google Scholar]
- 88.van der Laan MJ, Pollard KS. A new algorithm for hybrid hierarchical clustering with visualization and the bootstrap. J Stat Plan Inference. 2003;117(2):275–303. [Google Scholar]
- 89.Gardiner L-J, Quinton-Tulloch M, Olohan L, et al. A genome-wide survey of DNA methylation in hexaploid wheat. Genome Biol. 2015;16(1):273. doi: 10.1186/s13059-015-0838-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y, Harris CJ, Liu Q, et al. Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proc Natl Acad Sci U S A. 2018;115(5):E1069–74. doi: 10.1073/pnas.1716300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mager S, Ludewig U. Massive loss of DNA methylation in nitrogen-, but not in phosphorus-deficient Zea mays roots is poorly correlated with gene expression differences. Front Plant Sci. 2018;9:497. doi: 10.3389/fpls.2018.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lindroth AM, Cao X, Jackson JP, et al. Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science. 2001;292(5524):2077–80. doi: 10.1126/science.1059745. [DOI] [PubMed] [Google Scholar]
- 93.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–20. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barrero MJ, Boue S, Izpisua Belmonte JC. Epigenetic mechanisms that regulate cell identity. Cell Stem Cell. 2010;7(5):565–70. doi: 10.1016/j.stem.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 95.Juhling F, Kretzmer H, Bernhart SH, et al. Metilene: fast and sensitive calling of differentially methylated regions from bisulfite sequencing data. Genome Res. 2016;26(2):256–62. doi: 10.1101/gr.196394.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Catoni M, Tsang JM, Greco AP, et al. DMRcaller: a versatile R/bioconductor package for detection and visualization of differentially methylated regions in CpG and non-CpG contexts. Nucleic Acids Res. 2018;46(19):e114. doi: 10.1093/nar/gky602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hebestreit K, Dugas M, Klein HU. Detection of significantly differentially methylated regions in targeted bisulfite sequencing data. Bioinformatics. 2013;29(13):1647–53. doi: 10.1093/bioinformatics/btt263. [DOI] [PubMed] [Google Scholar]
- 98.Kurdyukov S, Bullock M. DNA methylation analysis: choosingthe right method. Biology (Basel) 2016;5(1):E3. doi: 10.3390/biology5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robinson MD, Kahraman A, Law CW, et al. Statistical methods for detecting differentially methylated loci and regions. Front Genet. 2014;5:324. doi: 10.3389/fgene.2014.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–22. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ashikawa I. Gene-associated CpG islands in plants as revealed by analyses of genomic sequences. Plant J. 2001;26(6):617–25. doi: 10.1046/j.1365-313x.2001.01062.x. [DOI] [PubMed] [Google Scholar]
- 102.Ashikawa I. Gene-associated CpG islands and the expression pattern of genes in rice. DNA Res. 2002;9(4):131–4. doi: 10.1093/dnares/9.4.131. [DOI] [PubMed] [Google Scholar]
- 103.Bock C, Reither S, Mikeska T, et al. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21(21):4067–8. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 104.Lin X, Sun D, Rodriguez B, et al. BSeQC: quality control of bisulfite sequencing experiments. Bioinformatics. 2013;29(24):3227–9. doi: 10.1093/bioinformatics/btt548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lukauskas S, Visintainer R, Sanguinetti G, et al. DGW: an exploratory data analysis tool for clustering and visualisation of epigenomic marks. BMC Bioinformatics. 2016;17(16):447. doi: 10.1186/s12859-016-1306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chari R, Thu KL, Wilson IM, et al. Integrating the multiple dimensions of genomic and epigenomic landscapes of cancer. Cancer Metastasis Rev. 2010;29(1):73–93. doi: 10.1007/s10555-010-9199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sunami E, de Maat M, Vu A, et al. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6(4):e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ji H, Zhou C, Pan R, et al. APOE hypermethylation is significantly associated with coronary heart disease in males. Gene. 2018;689:84–9. doi: 10.1016/j.gene.2018.11.088. [DOI] [PubMed] [Google Scholar]
- 109.Ghavifekr Fakhr M, Farshdousti Hagh M, Shanehbandi D, et al. DNA methylation pattern as important epigenetic criterion in cancer. Genet Res Int. 2013;2013 doi: 10.1155/2013/317569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Finnegan EJ, Genger RK, Kovac K, et al. DNA methylation and the promotion of flowering by vernalization. Proc Natl Acad Sci USA. 1998;95(10):5824–9. doi: 10.1073/pnas.95.10.5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eichten SR, Springer NM. Minimal evidence for consistent changes in maize DNA methylation patterns following environmental stress. Front Plant Sci. 2015;6:308. doi: 10.3389/fpls.2015.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li J, Huang Q, Sun M, et al. Global DNA methylation variations after short-term heat shock treatment in cultured microspores of Brassica napus cv. Sci Rep. 2016;6 doi: 10.1038/srep38401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun S, Noviski A, Yu X. MethyQA: a pipeline for bisulfite-treated methylation sequencing quality assessment. BMC Bioinformatics. 2013;14:259. doi: 10.1186/1471-2105-14-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Worm Orntoft MB, Jensen SO, Hansen TB, et al. Comparative analysis of 12 different kits for bisulfite conversion of circulating cell-free DNA. Epigenetics. 2017;12(8):626–36. doi: 10.1080/15592294.2017.1334024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Holmes EE, Jung M, Meller S, et al. Performance evaluation of kits forbisulfite-conversion of DNA from tissues, cell lines, FFPE tissues, aspirates, lavages, effusions, plasma, serum, and urine. PLoS One. 2014;9(4):e93933. doi: 10.1371/journal.pone.0093933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu YY, Cui HM. The method of estimating bisulfite conversion rate in DNA methylation analysis. Yi Chuan. 2015;37(9):939–44. doi: 10.16288/j.yczz.15-129. [DOI] [PubMed] [Google Scholar]
- 117.International Wheat Genome Sequencing Consortium. Appels R, Eversole K, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018;361(6403):eaar719. doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 118.Kawakatsu T, Huang SC, Jupe F, et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166(2):492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schmitz RJ, Schultz MD, Urich MA, et al. Patterns of population epigenomic diversity. Nature. 2013;495(7440):193–8. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hoffmann S, Otto C, Kurtz S, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol. 2009;5(9):e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tran H, Porter J, Sun MA, et al. Objective and comprehensive evaluation of bisulfite short read mapping tools. Adv Bioinformatics. 2014:2014. doi: 10.1155/2014/472045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lee JH, Park SJ, Kenta N. An integrative approach for efficient analysis of whole genome bisulfite sequencing data. BMC Genomics. 2015;16(12):S14. doi: 10.1186/1471-2164-16-S12-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hatem A, Bozdag D, Toland AE, et al. Benchmarking short sequence mapping tools. BMC Bioinformatics. 2013;14:184. doi: 10.1186/1471-2105-14-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Harris EY, et al. BRAT: bisulfite-treated reads analysis tool. Bioinformatics. 2010;26(4):572–3. doi: 10.1093/bioinformatics/btp706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dinh HQ, Dubin M, Sedlazeck FJ, et al. Advanced methylome analysis after bisulfite deep sequencing: an example in Arabidopsis. PLoS One. 2012;7(7):e41528. doi: 10.1371/journal.pone.0041528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chinnusamy V, Zhu JK. Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol. 2009;12(2):133–9. doi: 10.1016/j.pbi.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kumar S. Epigenomics of plant responses to environmental stress. Epigenomes. 2018;2(1):6. [Google Scholar]
- 128.Lämke J, Bäurle I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017;18(1):124. doi: 10.1186/s13059-017-1263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gutzat R, Mittelsten Scheid O. Epigenetic responses to stress: triple defense? Curr Opin Plant Biol. 2012;15(5):568–73. doi: 10.1016/j.pbi.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kinoshita T, Seki M. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 2014;55(11):1859–63. doi: 10.1093/pcp/pcu125. [DOI] [PubMed] [Google Scholar]
- 131.Yang Y, Guo Y. Unraveling salt stress signaling in plants. J Integr Plant Biol. 2018;60(9):796–804. doi: 10.1111/jipb.12689. [DOI] [PubMed] [Google Scholar]
- 132.Abhinandan K, Skori L, Stanic M, et al. Abiotic stress signaling in wheat—an inclusive overview of hormonal interactions during abiotic stress responses in wheat. Front Plant Sci. 2018;9:734. doi: 10.3389/fpls.2018.00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gallusci P, Hodgman C, Teyssier E, et al. DNA methylation and chromatin regulation during fleshy fruit development and ripening. Front Plant Sci. 2016;7:807. doi: 10.3389/fpls.2016.00807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Manning K, Tor M, Poole M, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38(8):948–52. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 135.Omidvar V, Fellner M. DNA methylation and transcriptomic changes in response to different lights and stresses in 7B-1 male-sterile tomato. PLoS One. 2015;10(4):e0121864. doi: 10.1371/journal.pone.0121864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Farinati S, Rasori A, Varotto S, et al. Rosaceae fruit development, ripening and post-harvest: an epigenetic perspective. Front Plant Sci. 2017;8:1247. doi: 10.3389/fpls.2017.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Boyko A, Kovalchuk I. Epigenetic control of plant stress response. Enuiron Mol Mutagen. 2008;49(1):61–72. doi: 10.1002/em.20347. [DOI] [PubMed] [Google Scholar]
- 138.White NR, Barfield RJ. Playback of female rat ultrasonic vocalizations during sexual behavior. Physiol Behau. 1989;45(2):229–33. doi: 10.1016/0031-9384(89)90123-6. [DOI] [PubMed] [Google Scholar]
- 139.Xu S, Chong K. Remembering winter through vernalisation. Nat Plants. 2018;4(12):997–1009. doi: 10.1038/s41477-018-0301-z. [DOI] [PubMed] [Google Scholar]
- 140.Ehrlich M, Gama-Sosa MA, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10(8):2709–21. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.He XJ, Chen T, Zhu JK. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011;21(3):442–65. doi: 10.1038/cr.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yi SV. Insights into epigenome evolution from animal and plant methylomes. Genome Biol Euol. 2017;9(11):3189–201. doi: 10.1093/gbe/evx203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Su Z, Han L, Zhao Z. Conservation and divergence of DNA methylation in eukaryotes: new insights from single base-resolution DNA methylomes. Epigenetics. 2011;6(2):134–40. doi: 10.4161/epi.6.2.13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lentini A, Lagerwall C, Vikingsson S, et al. A reassessment ofDNA-immunoprecipitation-based genomic profiling. Nat Methods. 2018;15(7):499–504. doi: 10.1038/s41592-018-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ziller MJ, Hansen KD, Meissner A, et al. Coverage recommendations for methylation analysis by whole-genome bisulfite sequencing. Nat Methods. 2015;12(3):230. doi: 10.1038/nmeth.3152. 232-2,1 p following. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li Q, Song J, West PT, et al. Examining the causes and consequences of context-specific differential DNA methylation in maize. Plant Physiol. 2015;168(4):1262–74. doi: 10.1104/pp.15.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.