Abstract
Objective
Coordination ellipsis is a linguistic phenomenon abound in medical text and is challenging for concept normalization because of difficulty in recognizing elliptical expressions referencing 2 or more entities accurately. To resolve this bottleneck, we aim to contribute a generalizable method to reconstruct concepts from medical coordinated elliptical expressions in a variety of biomedical corpora.
Materials and Methods
We proposed a graph-based representation model and built a pipeline to reconstruct concepts from coordinated elliptical expressions in medical text (RECEEM). There are 4 modules: (1) identify all possible candidate conjunct pairs from original coordinated elliptical expressions, (2) calculate coefficients for candidate conjuncts using the embedding model, (3) select the most appropriate decompositions by global optimization, and (4) rebuild concepts based on a pathfinding algorithm. We evaluated the pipeline’s performance on 2658 coordinated elliptical expressions from 3 different medical corpora (ie, biomedical literature, clinical narratives, and eligibility criteria from clinical trials). Precision, recall, and F1 score were calculated.
Results
The F1 scores for biomedical publications, clinical narratives, and research eligibility criteria were 0.862, 0.721, and 0.870, respectively. RECEEM outperformed 2 previously released methods. By incorporating RECEEM into 2 existing NLP tools, the F1 scores increased from 0.248 to 0.460 and from 0.287 to 0.630 on concept mapping of 1125 coordination ellipses.
Conclusions
RECEEM improves concept normalization for medical coordinated elliptical expressions in a variety of biomedical corpora. It outperformed existing methods and significantly enhanced the performance of 2 notable NLP systems for mapping coordination ellipses in the evaluation. The algorithm is open sourced online (https://github.com/chiyuan1126/RECEEM).
Keywords: coordination ellipsis, concept normalization, natural language processing
INTRODUCTION
A wealth of reusable medical information and knowledge is accumulating as free text in clinical narratives, biomedical literature, or clinical trials.1,2 Information extraction and knowledge engineering from these data hinges on accurate and efficient concept recognition and concept mapping to specific terminologies and ontologies. The latter task (concept normalization) is often significantly impaired by inaccurate concept recognition of coordinated elliptical expressions.3 In linguistics, ellipsis is a grammatical device that achieves textual concision by omitting repeated words. For instance, the phrase “breast and squamous cell neoplasms” expresses “breast neoplasms” and “squamous cell neoplasms” by sharing “neoplasms” between “breast” and “squamous cell.” Commonly used medical or clinical natural language processing (NLP) tools are capable of recognizing elliptical expressions as entities, but they have difficulty in mapping the expressions to the correct set of concepts. For example, CLAMP (Clinical Language Annotation, Modeling, and Processing) did not decompose the expression, mapping the whole expression to the concept “squamous cell neoplasms” and missing “breast neoplasm” (Table 1). cTAKES (clinical Text Analysis and Knowledge Extraction System) produced “breast” and “squamous cell neoplasms” instead of reconstructing “breast neoplasm,” resulting in partially inaccurate, misrepresented mappings. MetaMap with conjunction processing enabled maps the phrase to “breast neoplasms” and “squamous cell” (instead of “squamous cell neoplasms”). On the one hand, considering that specialized concepts are common in technical languages, coordination ellipses are more frequently used in medical language than in the general domain.3 On the other hand, more granular results are required for medical downstream tasks such as free-text based phenotyping and cohort identification.8 To tackle such mapping issues, current NLP pipelines mainly utilize complete or partial rule-based or ad hoc methods. Such methods lack generalizability to different types of biomedical text and may require extensive customization to design corpus-specific patterns.
Table 1.
Examples of a coordinated elliptical expression processed by widely used medical NLP tools
|
Text: “this highly conserved putative oncogene, which encodes a novel cyclin, has been linked to BCL1 and implicated also in subsets of breast and squamous cell neoplasms with 11q13 amplification.” (PMID : 1682919) | ||
|---|---|---|
| Method | Named Entity Recognition results | Concept Mapping Results |
| CLAMP4 |
|
squamous cell neoplasms (C0206720) |
| cTAKES5 |
|
|
| MetaMap6 (w/o--conj) | 760 C0006141: BREAST (Breast) [Body Part, Organ, or Organ Component] | |
| 833 C0206720: Cell Neoplasms, Squamous (Squamous Cell Neoplasms) [Neoplastic Process] | ||
| MetaMap7 (w--conj) | 598 C0221910: Squamous Cell (Squamous Epithelial Cells) [Cell] | |
| 773 C1458155: Neoplasm of Breast (Mammary Neoplasms) [Neoplastic Process] | ||
“MetaMap w/o –conj” refers to MetaMap without turning on conjunction processing; “MetaMap w–conj” refers to MetaMap with conjunction processing turned on.
CLAMP: Clinical Language Annotation, Modeling, and Processing; cTAKES: clinical Text Analysis and Knowledge Extraction System; NLP: natural language processing; RECEEM: reconstruct concepts from coordinated elliptical expressions in medical text.
To bridge this knowledge gap, we contributed an unsupervised method to reconstruct concepts from coordinated elliptical expressions in medical text (RECEEM) using a graph-based model. A large-scale phrase set (ie, PubMed Phrases)9 was used to train an embedding model using word2vec to measure parallelism between candidate conjuncts.10 All candidate conjuncts are enumerated, and the most appropriate decomposing conjuncts are selected based on parallelism. A generalizability evaluation was conducted on elliptical entities identified from 3 medical corpora: biomedical literature, clinical narratives, and eligibility criteria in clinical trials. RECEEM can be added to existing medical NLP tools to facilitate concept normalization of elliptical expression in medical texts. We released the benchmark data, source code, and pretrained model online (https://github.com/chiyuan1126/RECEEM).
Related work
Approaches to reconstruct conjunction ellipses in concept normalization can be pattern or rule-based, statistics-based, or hybrid methods. Different methods are conducted at different granularities from the phrase level to the sentence level. Among pattern-based methods for identifying conjuncts, Nhan et al11 designed a transformation component to parse verb phrase ellipses by expanding conjunctions and reconstructing full sentences by filling in gapped information. Okumura and Muraki12 proposed a symmetric pattern to expand English coordination structures for improving English-Japanese translation. Klavans and Jacquemin13 presented a corpus-based system to expand multiword index terms using linguistic part of speech rules and morphological analysis results. Goldberg14 proposed an unsupervised model for statistically determining prepositional phrase attachment; the model was trained with unannotated 1988 Wall Street Journal text and achieved 72% accuracy. Teranishi et al15 developed a deep neural network model incorporating 2 conjunct properties (similarity and replaceability), improving clause-level coordination identification. The model identifies the boundary of the entire coordinate structure, identifying the italicized section in the following example: “the tender offer for a combination of cash, Memotec stock and debentures.”
Coordination resolution research has also attracted attention in the biomedical domain. A conjunction resolving function was added to MetaMap 2016v2 (“–conj” configuration option), combining the enumeration method and the dictionary look-up method to recombine concepts from conjunctions.7 Buyko et al16 employed a conditional random field (CRF)–based method extending the feature set to extract named entities. Subsequently, coordinated compound entities are screened out according to a set of conjunctions generated by statistical results. Chae et al17 also adopted a CRF model and predefined forward, backward, and complex coordination ellipses and developed a pattern-based method using lexicons to identify regions of different components (ie, conjunction, conjuncts, ellipsis antecedent). Wei et al3 proposed SimConcept, integrating a CRF model with pattern identification in a pattern-abundant pipeline. SimConcept designed 4 patterns in token reassembly and 2 heuristic methods in postprocessing. Overall, SimConcept achieved high performance on 5 biological corpora for genes (BioCreative 2 GN task train/test corpus and National Library of Medicine GIA corpus), diseases (National Center for Biotechnology Information [NCBI] Disease corpus), and chemicals (BioCreative IV ChemDNER task corpus). Of note, SimConcept can parse ellipses within a word boundary; for example, “BRCA1/2” is parsed as “BRCA1” and “BRCA2,” “T1-4 breast cancer” is annotated as “T1 breast cancer,” “T2 breast cancer,” “T3 breast cancer,” and “T4 breast cancer.” In contrast, our method focuses exclusively on token level reconstruction and does not parse character-level ellipses or ellipses within a word boundary. Jiang18 trained a clinical syntax parser with a large clinical Treebank (MiPACQ) and integrated a rule-based method using semantic information to solve coordination ambiguity issues based on syntax parsing results. The relation between coordinated elliptical expressions and their attributes was disambiguated. However, the coordination disambiguation method was developed to solve the ambiguity in relation extraction and did not conduct ellipsis expansion for the original coordinated elliptical expressions. Blake Rindflesch19 employed syntactic dependencies to extract forward, backward and complex ellipses from PubMed literature. Dependencies were adopted in building a dictionary of noncoordinated noun phrases to test new generated phrases. Shimbo and Hara20 proposed a discriminative learning model that only required a small training set and minimal features to detect and disambiguate coordinated noun phrases in the GENIA corpus.
In contrast to the previously mentioned studies, RECEEM is an unsupervised approach to reconstructing concepts from coordinated elliptical expressions that generalizes well across biomedical corpora, obviating laborious corpora-specific pattern design. RECEEM operates directly on the entities identified by existing named entity recognition (NER) modules (example provided in the Materials and Methods), allowing it to be easily incorporated into established medical or clinical information extraction pipelines.
MATERIALS AND METHODS
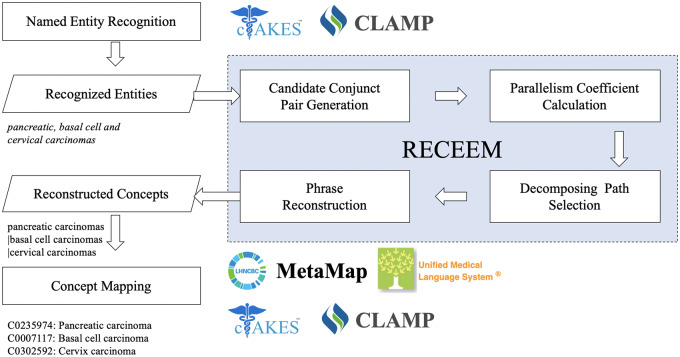
The RECEEM pipeline consists of 4 steps: candidate conjunct pair generation, parallelism coefficient calculation, decomposing path selection, and phrase reconstruction (Figure 1). The input includes entities with conjunctions identified from existing NLP systems’ NER modules. Similar to Buyko et al’s work,16 all NER output are considered, but entities without conjunctions are screened out. Then, any named entity containing conjunctions are decomposed and reconstructed into multiple entities. Continuing with the examples from Table 1, from CLAMP’s output (“breast and squamous cell neoplasm”), RECEEM detects the conjunction and decomposes the entity into “breast neoplasm” and “squamous cell neoplasm.” From cTAKES’ output (“breast” and “squamous cell neoplasm”), RECEEM processes each entity individually and returns both entities unaltered because neither contains a conjunction.
Figure 1.
RECEEM (reconstruct concepts from coordinated elliptical expressions in medical text) pipeline for reconstructing concepts from coordinated elliptical expressions.
Task statement
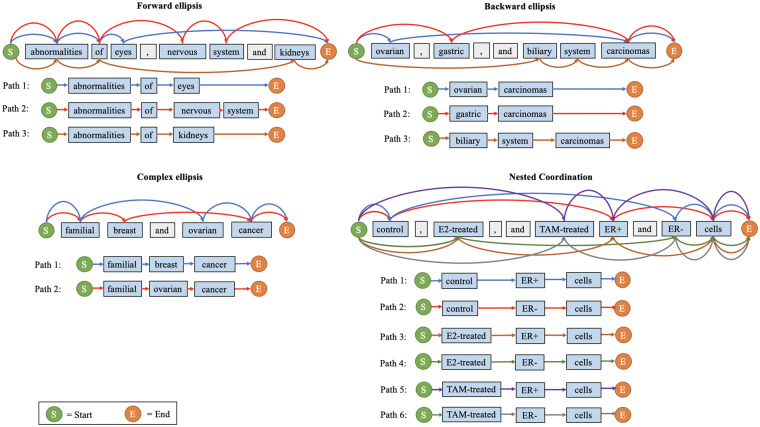
In general, coordination ellipses mainly have 4 categories17: forward ellipsis (eg, “abnormalities of eyes, nervous system, and kidneys”), backward ellipsis (eg, “breast and ovarian cancer”), complex ellipsis (eg, “familial breast and ovarian cancers”), and nested coordination (eg, “control, E2-treated, and TAM-treated ER+ and ER- cells”). Figure 2 illustrates these examples.
Figure 2.
A unified representation model for coordination resolution. Each distinct color identifies a resolved conjunct.
In this article, we use the following terminology consistent with previous works to describe coordinated elliptical expressions.16 A conjunction (eg, “,” [comma] and “and” in Figure 2) is a word or symbol that connects 2 or more conjuncts (eg, “eyes,” “nervous system,” “kidneys”) in the phrase. An antecedent (eg, “abnormalities of” and “carcinomas”) is the part of an elliptical mention shared by all conjuncts. The phrases reconstructed by combining the antecedent with each of the conjuncts are called resolved conjuncts. In forward, backward, and complex ellipses, a conjunct does not include the antecedent, but in nested coordination, each word could have more than 1 role. For example, in the nested elliptical expression “control, E2-treated, and TAM-treated ER+ and ER- cells,” “control” and “ER+” (among others) can act as antecedent or conjunct for different resolved conjuncts.
To build a unified processing pipeline for all 4 categories of coordinated elliptical expressions, we propose a graph-based representation model. All words and symbols in the expression are vertices sequentially connected in their original order (Figure 2). We add a starting vertex before the first word and an ending vertex after the last word. The edges between vertices are updated according to the “parallelism principle, which states that all conjuncts in an elliptical expression must exhibit parallelism with each other.21 In our representation model, all paths from starting vertex to ending vertex are resolved conjuncts. Using this graph representation model, the coordination resolution problem is transformed into a path-finding problem. The details of our method are described subsequently.
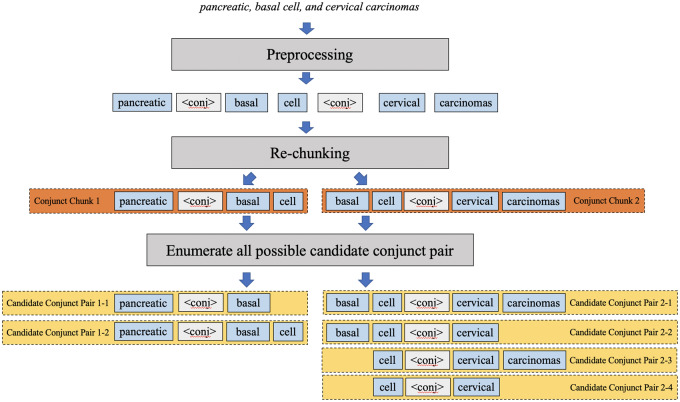
Candidate conjunct pair generation
The key task in reconstructing concepts from coordinated elliptical expressions is to determine the boundaries of conjuncts. Coordinated elliptical expressions can contain more than 1 conjunction, such as “, (comma), (comma) and” in the phrase “pancreatic, basal cell, and cervical carcinomas.” To unify the process for various ellipses, we tackled conjunctions individually, allowing nested coordination and multiconjunction cases to be resolved with the same pipeline. A candidate conjunct pair generation module was designed to enumerate candidate conjunct pairs from coordinated elliptical expressions (Figure 3). First, elliptical expressions are tokenized. All conjunctions are identified and replaced by a uniform conjunction mark <conj>, with multiple continuous conjunctions converted to a single conjunction mark. Then, rechunking is conducted to split the original expression into multiple conjunct chunks such that each conjunct chunk only keeps 1 conjunction mark. In each conjunct chunk, all possible conjuncts are enumerated. For example, “pancreatic <conj> basal cell” is decomposed into “pancreatic vs. basal” and “pancreatic vs. basal cell” for the following module to select the most appropriate conjunct pair by comparing parallelism.
Figure 3.
An example of candidate conjunct pair generation.
Parallelism coefficient calculation
Given the parallel structure of coordinated elliptical expressions,21 quantitative measurements of parallelism between candidate conjuncts is critical for determining the most appropriate conjunct pairs for entity decomposition and reconstruction. Correspondingly, we propose an embedding model to calculate the parallelism coefficient which reflects the parallelism among candidate conjuncts.
Parallelism coefficient definition
In all 4 forms of coordination ellipses, conjuncts are formed by continuous sequences of words, and conjunct resolution always proceeds by combining antecedents and conjuncts in a forward-moving direction. Hence, potential complemented concepts are enumerated unidirectionally, evaluating continuous blocks of words for each candidate conjunct (Figure 3). Specifically, the parallelism coefficient of a candidate conjunct pair is defined in equation 1.
| Eq. 1 |
Here, is the index for <conj> in the chunk; is the candidate conjunct starting from index to ; and is the conjunct candidate starting from index to . The range of is from 0 to and the range of is from to the last index of the conjunct chunk. The complexity of generating all possible candidate conjunct pairs is C(n*m), where n is the number of tokens ahead of the conjunction and m is the number of tokens after the conjunction. The parallelism is measured by the absolute value of the cosine distance between the vectors of the 2 conjuncts generated from our embedding model, described below.
Embedding model for measuring parallelism coefficient
To measure the parallelism between candidate conjuncts in each candidate conjunct pair, a concept embedding model was trained with all PubMed abstracts published before November 2017. To include a large phrase vocabulary, we employed an open set of coherent medical phrases—PubMed Phrase,9 including all phrase types (eg, adjective, prepositional, or noun phrases). PubMed Phrase contains 705 915 phrases and was collected by the hypergeometric test and filtered by the BM25 ranking function. The efficient Aho–Corasick string-searching algorithm22 was used as a fast pattern-matching method to find instances of these biomedical phrases in the abstracts. Multiword expressions were formatted as hyphen-connected words (eg, word1-word2-…-wordi) in the embedding model’s training corpus. With hyphenation, the training model treats a multiword concept like a single word concept, allowing comparison of single and multiword concepts in parallelism coefficient measurement. Additionally, the hyphenation expands the effective vocabulary from 1 828 643 to 2 489 984 terms. We utilized the word2vec algorithm to prepare the PubMed phrase2vec model. A Skip-Gram model with 5-length window size was configured for the PubMed phrase2vec training. The PubMed phrase2vec model was used as a dictionary to support phrase vector lookup in calculating parallelism coefficients. If some candidate conjunct is not in the expanded vocabularies, the coefficient for the relevant phrase will be zero. For example, “II diabetes” from “Type I or II diabetes mellitus,” is not in the vocabularies, so the parallelism coefficient between “I” and “II diabetes” is zero.
Decomposing path selection
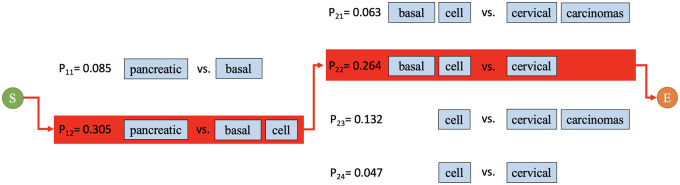
When elliptical entity expressions have a single conjunction, selecting the optimal candidate conjunct pair is trivial. All candidate conjunct pairs are enumerated along with their parallelism coefficients, and the pair with the greatest parallelism is selected to expand with the antecedent. For example, in “breast and squamous cell neoplasms, the parallelism coefficient between “breast” and “squamous cell” is greater than that between “breast” and “squamous” or “breast” and “squamous cell neoplasms,” thus “breast” and “squamous cell” are selected.
However, when multiple conjunctions are present, multiple paths through the graph model exist (Figure 4), and more than 1 decomposed result should be included during resolution of the entire coordinated elliptical expression. We reuse the parallelism coefficient calculated for each local candidate conjunct pair for global optimization of the decomposition. We defined an overall decomposing probability (P) to evaluate overall decomposing performance according to the parallelism principle (equation 2). The path with the maximum overall decomposition probability is chosen to optimize overall performance and balance the loss and gain in each subdecomposition.
| Eq. 2 |
Figure 4.
An example of decomposing path selection. All possible paths between the start and end vertices are evaluated. The algorithm selects the path with the greatest overall decomposition probability (equation 2). In this example, the path including “pancreatic, “basal cell,” and “cervical” yields the greatest overall decomposition probability.
Phrase reconstruction
After identifying the globally optimal decomposed path, we reassemble the words to resolve the conjuncts in accordance with the graph-based model. The phrase reconstruction module comprises 3 steps: graph initialization, edge update, and path finding (Figure 5 and Supplementary Appendix 1.1). The graph is initialized as previously described (see Task Statement). The following edge update procedures are executed in every conjunct chunk in sequence. Within each conjunct chunk, the original incoming and outgoing edge to and from <conj> will be updated as the selected candidate conjunct pair. The edges with the source vertex of <conj> will be updated to start from the same source vertex in the first word of the first conjunct. The edges with the target vertex of <conj> will be redirected to the same target vertex in the second word of the second conjunct. In the path finding step, all paths from “<Start>” to “<End>” are followed to generate the resolved conjuncts.
Figure 5.
Phrase reconstruction comprises 3 steps: (1) graph initialization, (2) edge update in accordance with the globally optimal decomposition, and (3) path finding to generate all resolved conjuncts.
Evaluation methods
Evaluation data and metrics
Our evaluation experiments were performed on 3 types of medical corpora: biomedical literature, eligibility criteria from ClinicalTrials.gov, and clinical narratives. Specifically, the biomedical literature data were from the NCBI Disease corpus23 and GENIA dataset.24 The gold standard reconstructed concepts for these projects were already well annotated. Eligibility criteria data were from the CHIA dataset’s8 gold standards and from NER results from 1000 additional trials using an NER model trained with CHIA data. Clinical narratives data were collected from the 2010 i2b2 Challenge.25 Two annotators with clinical backgrounds provided gold standard reconstructed results for clinical narratives and eligibility criteria data above their original annotated NER data. The annotators first made independent annotations, then met and discussed discrepancies, revised the annotation guideline as needed, and iterated these processes until full consensus was reached. After removing duplicate entities, there were 2658 coordinated elliptical expressions in total, with 1553, 126, and 979 coordinated elliptical expressions coming from biomedical literature, clinical narratives, and eligibility criteria, respectively. RECEEM was evaluated based on the number of true positives, false negatives, and false positives in the reconstructed results. Exact matches between reconstructed entities generated by RECEEM and the gold standards were counted as true positives. False negatives represented reconstructed entities provided by the gold standard but not predicted by RECEEM. False positives represented reconstructed entities predicted by RECEEM but not provided by the gold standard. Precision, recall, and F1 measure were calculated for evaluating the overall performance.
Comparison of different pretrained models and related systems
We evaluated alternative pretrained NLP models for calculating the parallelism coefficient. We tested our pipeline integrated with other pretrained language models—contextualized model BERT,26 BioBERT,27 ClinicalBERT,28 and phrase2vec.29 We calculated the 95% confidence intervals (CIs) for all performance metrics using the adjusted bootstrap percentile interval with 10 000 iterations. Statistical significance tests were calculated from 10 000 iterations of paired bootstrap30 on the combined dataset, sampling with replicates and calculating P value as the percentage of iterations with positive performance difference.
We also performed these evaluations using MetaMap and SimConcept for comparison. We employed MetaMap version 2018v2 configured to process conjunctions. MetaMap’s conjunction processing does not attempt to identify a consistent set of reconstructed concepts, but rather outputs a list of mapping candidates. To make MetaMap’s results comparable with ours and reduce its false positives, we manually reviewed MetaMap’s results and removed duplicated and subconcept mappings. SimConcept was run on “mentions input” mode and was employed with its well-trained model and predefined patterns.
Enhancement for current NLP tools
To evaluate the effectiveness of RECEEM on augmenting existing medical NLP tools, we applied RECEEM in 2 widely used NLP tools: CLAMP4 and Apache cTAKES.5 CLAMP is a toolkit for efficiently building customized clinical NLP pipelines.4 cTAKES is an NLP system for extracting information from clinical texts.5 The DF_Dictionary_based_UMLS_encoder in CLAMP’s concept mapping module was employed during concept normalization. One domain expert manually mapped all medical free-text concepts from the i2b2, NCBI Disease, and CHIA datasets to Unified Medical Language System concept unique identifiers, which were used as gold standards. After removing the duplicates and unmapped terms, there were 1125 gold standard concepts in total.
RESULTS
Comparison of pretrained models
Table 2 shows the performance results comparing different pretrained models for calculating parallelism. RECEEM incorporating the PubMed phrase2vec model earned the highest performance, achieving an F1 score of 0.859 on all combined corpora, 0.862 for biomedical literature, 0.721 for clinical narratives, and 0.870 for eligibility criteria. The phrase2vec-based models outperformed the BERT-based models with statistical significance. On the combined corpora, PubMed phrase2vec and phrase2vec achieved F1 scores of 0.859 (95% CI: 0.845-0.872) and 0.850 (95% CI, 0.836-0.863), respectively, whereas BioBERT, ClinicalBERT, and BERT achieved 0.752 (95% CI, 0.737-0.767), 0.751 (95% CI, 0.736-0.766), and 0.750 (95% CI, 0.735-0.766), respectively. From the paired bootstrap test results, PubMed phrase2vec’s F1 scores were significantly better than phrase2vec’s (P < .05), with an average improvement of 0.0092.
Table 2.
Results of RECEEM integrated with different pretrained language models
| Biomedical literature (n = 1553) |
Clinical narrative (n = 126) |
Eligibility criteria (n = 979) |
Total (N = 2658) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 | Precision | Recall | F1 |
| RECEEM (BERT) | 0.777 | 0.781 | 0.779 | 0.667 | 0.681 | 0.674 | 0.716 | 0.727 | 0.721 | 0.747 | 0.754 | 0.750 |
| 2641/3398 | 2641/3381 | 0.762-0.796 | 196/294 | 196/288 | 0.600-0.748 | 1852/2586 | 1852/2549 | 0.694-0.748 | 4689/6278 | 4689/6218 | 0.735-0.766 | |
| 0.759-0.794 | 0.764-0.798 | 0.591-0.744 | 0.608-0.752 | 0.688-0.744 | 0.700-0.753 | 0.731-0.762 | 0.739-0.769 | |||||
| RECEEM (ClinicalBERT) | 0.741 | 0.746 | 0.743 | 0.695 | 0.712 | 0.703 | 0.761 | 0.774 | 0.767 | 0.747 | 0.756 | 0.751 |
| 2521/3404 | 2521/3381 | 0.726-0.761 | 205/295 | 205/288 | 0.627-0.779 | 1973/2594 | 1973/2549 | 0.741-0.794 | 4699/6293 | 4699/6218 | 0.736-0.766 | |
| 0.722-0.759 | 0.728-0.763 | 0.616-0.774 | 0.637-0.786 | 0.733-0.788 | 0.748-0.799 | 0.731-0.762 | 0.741-0.770 | |||||
| RECEEM (BioBERT) | 0.752 | 0.759 | 0.756 | 0.703 | 0.715 | 0.709 | 0.745 | 0.759 | 0.752 | 0.747 | 0.757 | 0.752 |
| 2566/3411 | 2566/3381 | 0.737-0.774 | 206/293 0.629-0.78 | 206/288 | 0.635-0.784 | 1935/2597 | 1935/2549 | 0.725-0.778 | 4707/6301 | 4707/6218 | 0.737-0.767 | |
| 0.733-0.771 | 0.741-0.777 | 0.642-0.789 | 0.717-0.772 | 0.733-0.785 | 0.731-0.762 | 0.742-0.771 | ||||||
| RECEEM (phrase2vec) | 0.846 | 0.855 | 0.850 | 0.699 | 0.708 | 0.703 | 0.854 | 0.876 | 0.865 | 0.842 | 0.857 | 0.850 |
| 2890/3418 | 2890/3381 | 0.834-0.867 | 204/293 | 204/288 | 0.623-0.783 | 2234/2615 | 2234/2549 | 0.841-0.888 | 5328/6329 | 5328/6218 | 0.836-0.863 | |
| 0.827-0.864 | 0.839-0.870 | 0.616-0.780 | 0.626-0.787 | 0.828-0.880 | 0.854-0.897 | 0.827-0.857 | 0.844-0.870 | |||||
| RECEEM (PubMed phrase2vec) | 0.858 | 0.866 | 0.862a | 0.716 | 0.726 | 0.721a | 0.859 | 0.881 | 0.870a | 0.852 | 0.866 | 0.859a |
| 2929/3413 | 2929/3381 | 0.846-0.878 | 209/292 | 209/288 | 0.642-0.799 | 2245/2612 | 2245/2549 | 0.847-0.892 | 5383/6321 | 5383/6218 | 0.845-0.872 | |
| 0.841-0.875 | 0.851-0.881 | 0.635-0.795 | 0.645-0.804 | 0.833-0.884 | 0.859-0.901 | 0.837-0.867 | 0.853-0.878 | |||||
Values are n/n or 95% confidence interval, unless otherwise indicated.
RECEEM: reconstruct concepts from coordinated elliptical expressions in medical text.
The best performing result in the respective task.
Comparison with related systems
We compared our pipeline against SimConcept3 and MetaMap.6 As shown in Table 3, RECEEM achieved F1 measures of 0.862 (95% CI, 0.846-0.878), 0.721 (95% CI, 0.642-0.799), 0.870 (95% CI, 0.847-0.892), and 0.859 (95% CI, 0.845-0.8729 on the biomedical literature, clinical narrative, eligibility criteria, and combined corpora, respectively. Compared with SimConcept (0.142 [95% CI, 0.127-0.157], 0.379 [95% CI, 0.313-0.447], 0.427 [95% CI, 0.396-0.457], 0.266 [95% CI, 0.250-0.284]) and MetaMap (0.141 [95% CI, 0.128-0.155], 0.119 [95% CI, 0.08-0.164], 0.328 [95% CI, 0.304-0.353], 0.211 [95% CI, 0.198-0.224]), RECEEM performed significantly better on all tests.
Table 3.
Results of RECEEM and previously released methods
| Biomedical literature (n = 1553) |
Clinical narrative (n = 126) |
Eligibility criteria (n = 979) |
Total (N = 2658) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Method | Precision | Recal | F1 | Precision | Recal | F1 | Precision | Recal | F1 | Precision | Recal | F1 |
| MetaMap18v2 (conj++) | 0.124 | 0.164 | 0.141 | 0.104 | 0.139 | 0.119 | 0.322 | 0.334 | 0.328 | 0.193 | 0.232 | 0.211 |
| 553/4448 | 553/3381 | 0.128-0.155 | 40/385 | 40/288 | 0.08-0.164 | 852/2644 | 852/2549 | 0.304-0.353 | 1445/7477 | 1445/6218 | 0.198-0.224 | |
| 0.112-0.137 | 0.149-0.179 | 0.069-0.146 | 0.096-0.188 | 0.298-0.348 | 0.309-0.36 | 0.181-0.206 | 0.219-0.246 | |||||
| SimConcept | 0.154 | 0.132 | 0.142 | 0.423 | 0.344 | 0.379 | 0.490 | 0.378 | 0.427 | 0.296 | 0.242 | 0.266 |
| 445/2896 | 445/3381 | 0.127-0.157 | 99/234 | 99/288 | 0.313-0.447 | 963/1966 | 963/2549 | 0.396-0.457 | 1507/5096 | 1507/6218 | 0.250-0.284 | |
| 0.137-0.170 | 0.117-0.146 | 0.352-0.496 | 0.281-0.410 | 0.457-0.523 | 0.348-0.408 | 0.278-0.314 | 0.227-0.259 | |||||
| RECEEM (PubMed phrase2vec) | 0.858 | 0.866 | 0.862a | 0.716 | 0.726 | 0.721a | 0.859 | 0.881 | 0.870a | 0.852 | 0.866 | 0.859a |
| 2929/3413 | 2929/3381 | 0.846-0.878 | 209/292 | 209/288 | 0.642-0.799 | 2245/2612 | 2245/2549 | 0.847-0.892 | 5383/6321 | 5383/6218 | 0.845-0.872 | |
| 0.841-0.875 | 0.851-0.881 | 0.635-0.795 | 0.645-0.804 | 0.833-0.884 | 0.859-0.901 | 0.837-0.867 | 0.853-0.878 | |||||
Values are n/n or 95% confidence interval, unless otherwise indicated.
RECEEM: reconstruct concepts from coordinated elliptical expressions in medical text.
The best performing result in the respective task.
Enhancement for current NLP tools
We compared the performance of CLAMP and cTAKES stand-alone against their performance with RECEEM (Table 4). With the boost from RECEEM, cTAKES and CLAMP achieved higher F1 performances of 0.460 (95% CI, 0.436-0.484) and 0.630 (95% CI, 0.605-0.655), respectively, with a significant improvement over their stand-alone versions (0.248 [95% CI, 0.232-0.264] and 0.287 [95% CI, 0.269-0.305], respectively).
Table 4.
Concept normalization performance of existing NLP pipelines stand-alone and combined with RECEEM
| Methods | Precision | Recall | F1 |
|---|---|---|---|
| CLAMP | 0.477(597/1251) | 0.205(597/2906) | 0.287 |
| 0.449-0.505 | 0.191-0.22 | 0.269-0.305 | |
| CLAMP+RECEEM | 0.687(1693/2465) | 0.583(1693/2906) | 0.630a |
| 0.661-0.712 | 0.555-0.609 | 0.605-0.655 | |
| cTAKES | 0.286(636/2226) | 0.219(636/2906) | 0.248 |
| 0.266-0.305 | 0.204-0.233 | 0.232-0.264 | |
| cTAKES + RECEEM | 0.469(1312/2800) | 0.451(1312/2906) | 0.460a |
| 0.444-0.492 | 0.426-0.477 | 0.436-0.484 |
CLAMP: Clinical Language Annotation, Modeling, and Processing; cTAKES: clinical Text Analysis and Knowledge Extraction System; RECEEM: reconstruct concepts from coordinated elliptical expressions in medical text.
The best performing result in the respective task.
DISCUSSION
In this article, we presented RECEEM, a graph-based method to decompose coordinated elliptical expressions and construct nonelliptical concepts from them. By employing word embedding models to measure parallelism between candidate conjuncts, RECEEM performed well resolving composite mentions with ellipses in medical NLP without requiring handcrafted pattern designs or supervised annotations. In this section, a detailed error analysis of our results is discussed. Additionally, limitations and corresponding future improvements are outlined.
Error analysis
The task-specific evaluations (F1 = 0.859) indicate that our methods still have room for improvement. We performed an error analysis on all imperfect reconstructions of elliptical expressions, including partially inaccurate reconstructions. There were 516 imperfect reconstructions total. 19.2% (n = 99 of 516) of errors were caused by complex ellipses types that are out of the scope of our pipeline. For instance, some ellipses occur within a word boundary, which requires reconstruction on the character level, such as “hypo or hyperglycemia” which should be normalized to “hypoglycemia” and “hyperglycemia.” A total of 39.3% (n = 203 of 516) of errors were mainly due to poor results from the parallelism coefficient calculation generated by the pretrained model. For example, in “general and regional anesthesia, the cosine distance between the “general” and “regional anesthesia” vectors was higher than that between the “general” and “regional” vectors, hence an inaccurate reconstruction was generated. A total of 27.9% (n = 144 of 516) of errors were caused by the modifiers used in candidate conjuncts, which presents a challenge in parallelism comparison by embedding methods because the modified forms are not captured in the embedded vocabularies. For example, relevant phrases of a medical problem with body location are “left temporal lobe” and “right frontal lobe.” The remaining 13.6% (n = 70 of 516) of the errors occurred because of the incomplete vocabulary of our pretrained model. For example, “biliary system” was not included in the dictionary, resulting in a parallelism coefficient of 0 between “gastric” and “biliary system.” In these cases, PubMed phrase2vec provided no advantage for discriminating decomposing boundaries.
Limitations and future work
As an unsupervised method for decomposing and reconstructing medical coordination ellipses, RECEEM has several limitations. According to these limitations, we outline corresponding improvements for future studies.
First, the parallelism coefficient calculation method can be improved. Currently, the parallelism coefficient calculation relies heavily on the quality of our embedding model. In our study, PubMed phrase2vec worked well for most elliptical expressions with clearly defined meanings. However, parallelism calculations did not perform well for general terms such as “other” and “other organ.” For example, in the decomposition of “cardiac, pulmonary, hepatic, or other organ dysfunction,” the parallelism coefficients of “hepatic vs. other” and “hepatic vs. other organ” were both low, but “hepatic vs. other” was higher, leading to an incorrect decomposition. Moreover, modifiers in conjunct candidates may cause parallelism comparison to fail. For example, “susceptibility artifact in the left temporal lobe, right frontal lobe, and the splenius of the corpus callosum” has the modifiers, “left temporal,” “right frontal,” and “splenius of.” More advanced methods for comparing the parallelism of “left temporal lobe,” “right frontal lobe,” and “the splenius of the corpus callosum” are required. In addition, the incomplete vocabulary of the embedding model may also result in the failure of parallelism comparison. A potential resolution is to assign more weight in syntactic parsing results,19 enlarge the vocabulary used in the pretrained model, and improve the representation of the embedding model for adjectives.31
Second, possible gaps exist in the reconstruction process and medical terminologies standardization. Sometimes, a reconstruction that appears correct from a linguistic perspective can potentially harm subsequent concept normalization or medical dictionary look-up processes. Taking “waxing and waning sensorium” as an example, “waxing” and “waning” with disorders are stored in biomedical terminologies as entire concepts. In some cases, the entire elliptical coordinated expression is a proper disease name, such as “hand, foot and mouth disease,” and should not be decomposed into “hand disease,” “foot disease,” and “mouth disease.” A dictionary look-up model could be integrated with our current pipeline to check if the original phrase exists in the target terminologies. If the entire coordinated elliptical expression is an existing concept, reconstruction can be skipped.
In addition, the decomposition method is conducted on the token level and cannot handle character-level ellipses or ellipses within a word. In some numeric-related ellipses, a range of minimum and maximum numbers are adapted to avoid listing all the intermediate numbers. For example, “T1-4 breast cancer” should be reconstructed to “T1 breast cancer,” “T2 breast cancer,” “T3 breast cancer” and “T4 breast cancer.” On the other hand, some long words with the same suffix or postfix may have the ellipsis within the word. For instance, “hypo or hyperglycemia” should be recombined to “hypoglycemia” or “hyperglycemia.” The numeric ellipsis issue could be potentially solved by a pattern-based recognition model that is described in the SimConcept study.3 The character level ellipsis issue requires a vocabulary of common suffixes and postfixes, which can be generated using a clustering algorithm to execute against existing medical terminologies and vocabularies in the future.
Finally, our pure unsupervised method still has some shortcomings. No detailed patterns for coordination rules (eg, “prefix” + “A, B, C and D” or “A and B” + postfix) are used in our pipeline. As we described in the error analysis, the unsupervised method has some disadvantages when comparing modified phrases. This project is an early exploration of unsupervised methods for medical ellipsis reconstruction. More robust models may benefit from integrating diversified medical terminologies or dictionaries. A hybrid method combining our methods and inductive patterns from Wei et al’s work3 may perform better than our stand-alone methods, and is worth investigating in the future.
CONCLUSION
RECEEM is an early successful attempt to adopt pure unsupervised methods for resolving composite mentions with ellipses in medical text and does not require predefined patterns or training data. RECEEM can be generalized to different types of medical text and integrated with existing NLP pipelines to benefit end-to-end information extraction tasks.
FUNDING
This research was supported by the National Natural Science Foundation of China (nos. 61170035 and 61941113) (to YW), Fundamental Research Fund for the Central Universities (nos. 30918015103 and 30918012204) (to YW), and Nanjing Science and Technology Development Plan Project (no. 201805036) (to YW).
AUTHOR CONTRIBUTIONS
CY and YW conceived the methodology design together. CY designed and implemented the method. NS and CW helped identify and define the problem, discussed solutions, and edited the manuscript critically. CY, ZL, and RZ contributed to the design and evaluation of the system. All authors edited and approved the manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Xiangwei Guo from Dr. Wang’s lab for his kind help in data cleaning and data preprocessing. We also thank Alex Butler and Casey Ta from Dr. Weng’s lab for their help with the name selection for our tool and for editing the manuscript.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Luo J, Wu M, Gopukumar D, Zhao Y. Big data application in biomedical research and health care: a literature review. Biomed Inform Insights 2016; 8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meystre SM, Lovis C, Bürkle T, Tognola G, Budrionis A, Lehmann CU. Clinical data reuse or secondary use: current status and potential future progress. Yearb Med Inform 2017; 26 (1): 38–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wei C, Leaman R, Lu Z. SimConcept: a hybrid approach for simplifying composite named entities in biomedical text. IEEE J Biomed Health Inform 2015; 19 (4): 1385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soysal E, Wang J, Jiang M, et al. CLAMP—a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc 2018; 25 (3): 331–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 17 (5): 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001; 17–21. [PMC free article] [PubMed] [Google Scholar]
- 7. MetaMap. Processing conjuncts with --conj. https://metamap.nlm.nih.gov/Docs/FAQ/Conjunction.pdf Accessed June 13, 2019.
- 8. Kury FS, Fu L-H, Yuan C, Sim I, Carini S, Weng C. Hidden gaps in using common data models to achieve interoperability between electronic phenotypes and clinical data. In: AMIA 2019 Annual Symposium; November 18, 2019; Washington, DC.
- 9. Kim S, Yeganova L, Comeau DC, Wilbur WJ, Lu Z. PubMed Phrases, an open set of coherent phrases for searching biomedical literature. Sci Data 2018; 5: 180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blake G, Bly RW. The Elements of Technical Writing. New York, NY: Macmillan; 1993. [Google Scholar]
- 11. Nhan NT, Sager M, Lyman M, Tick LJ, Borst F, Su Y. A medical language processor for two Indo-European languages. Proc Annu Symp Comput Appl Med Care 1989; 554–8. [Google Scholar]
- 12. Okumura A, Muraki K. Symmetric pattern matching analysis for English coordinate structures. In: proceedings of the Fourth Conference on Applied Natural Language Processing; 1994: 41–6.
- 13. Klavans J., Jacquemin C. A natural language approach to multi-word term conflation. In: proceedings of the DELOS conference; 1997. https://www.ercim.eu/publication/ws-proceedings/DELOS3/Jacquemin.pdf. Accessed April 11, 2019.
- 14. Goldberg M. An unsupervised model for statistically determining coordinate phrase attachment. In: proceedings of the 37th Annual Meeting of the Association for Computational Linguistics on Computational Linguistics; 1999: 610–4.
- 15. Teranishi H, Shindo H, Matsumoto Y. Coordination boundary identification with similarity and replaceability. In: proceedings of the Eighth International Joint Conference on Natural Language Processing (Volume 1: Long Papers); 2017: 264–72.
- 16. Buyko E, Tomanek K, Hahn U. Resolution of coordination ellipses in biological named entities using conditional random fields. In: proceedings of the 10th Conference of the Pacific Association for Computational Linguistics; 2007: 163–71.
- 17. Chae J, Jung Y, Lee T, et al. Identifying non-elliptical entity mentions in a coordinated NP with ellipses. J Biomed Inform 2014; 47: 139–52. [DOI] [PubMed] [Google Scholar]
- 18. Jiang M. Improving Syntactic Parsing of Clinical Text Using Domain Knowledge [dissertation]. Houston, Texas, University of Texas Health Science Center at Houston, School of Biomedical Informatics; 2017.
- 19. Blake C, Rindflesch T. Leveraging syntax to better capture the semantics of elliptical coordinated compound noun phrases. J Biomed Inform 2017; 72: 120–31. [DOI] [PubMed] [Google Scholar]
- 20. Shimbo M, Hara K. A discriminative learning model for coordinate conjunctions. In: proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning (EMNLP-CoNLL); 2007: 610–9.
- 21. De Beaugrande R, Dressler WU. Introduction to Text Linguistics. Abingdon, United Kingdom: Routledge; 1981. [Google Scholar]
- 22.hankcs/AhoCorasickDoubleArrayTrie: An extremely fast implementation of Aho Corasick algorithm based on Double Array Trie. https://github.com/hankcs/AhoCorasickDoubleArrayTrie Accessed May 4, 2019.
- 23. Dogan RI, Leaman R, Lu Z. NCBI disease corpus: a resource for disease name recognition and concept normalization. J Biomed Inform 2014; 47: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim J-D, Ohta T, Tateisi Y, Tsujii J. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics 2003; 19 (suppl_1): i180–2. [DOI] [PubMed] [Google Scholar]
- 25. Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 18 (5): 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Devlin J,, Chang M-W,, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv:1810.04805v2; 2018.
- 27. Lee J, Yoon W, Kim S, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. arXiv:1901.08746v4; 2019. [DOI] [PMC free article] [PubMed]
- 28. Alsentzer E, Murphy JR, Boag W, et al. Publicly Available Clinical BERT Embeddings. arXiv:1904.03323v3; 2019.
- 29. Mikolov T, Sutskever I, Chen K, Dean J. Distributed representations of words and phrases and their compositionality. In: proceedings of Advances in Neural Information Processing Systems 26 (NIPS 2013); 2013: 3111–9.
- 30. Yeh A. More accurate tests for the statistical significance of result differences. In: proceedings of the 18th conference on Computational Linguistics. 2000; 2: 947–53.
- 31. Schwartz R, Reichart R, Rappoport A. Symmetric patterns and coordinations: fast and enhanced representations of verbs and adjectives. In: proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; 2016: 499–505.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.