Abstract
About 44.4 million people have been diagnosed with dementia worldwide, and it is estimated that this number will be almost tripled by 2050. Predicting mild cognitive impairment (MCI), an intermediate state between normal cognition and dementia and an important risk factor for the development of dementia is crucial in aging populations. MCI is formally determined by health professionals through a comprehensive cognitive evaluation, together with a clinical examination, medical history and often the input of an informant (an individual that know the patient very well). However, this is not routinely performed in primary care visits, and could result in a significant delay in diagnosis. In this study, we used deep learning and machine learning techniques to predict the progression from cognitively unimpaired to MCI and also to analyze the potential for patient clustering using routinely-collected electronic health records (EHRs). Our analysis of EHRs indicates that temporal characteristics of patient data incorporated in a deep learning model provides increased power in predicting MCI.
Keywords: mild cognitive impairment, dementia, Alzheimer’s disease, deep learning, machine learning, recurrent neural networks
I. Introduction
Dementia is one of the most prevalent health problems in the aging population. It is estimated that by 2030, 75.6 million people will suffer from various types of dementia worldwide, and that this number will increase to 135.5 million people in 2050 [1]. Such an increase will place a tremendous burden on patients, their families, society, and health care systems. Given that people with mild cognitive impairment (MCI) are at an increased risk for dementia, predicting MCI risk and understanding the progression from cognitively unimpaired (CU) to MCI and dementia is a crucial task to help the aging population with their health needs.
In general, MCI is formally determined by health professionals through a comprehensive cognitive evaluation, together with clinical examination, medical history and often the input of an informant (an individual that know the patient very well) to understand changes in cognition and daily function. However, this is not routinely performed in many primary care visits which results in a delay of timely diagnosis, misses opportunities for appropriate care plans, and leads to adverse clinical outcomes. Electronic health records (EHRs), especially clinical free text, contain valuable information that is routinely recorded as part of clinical care. This rich information may be used to identify patterns predicting the development of MCI and dementia. Previous studies have shown that some signals of cognitive decline exist in EHRs, years before the clinician diagnoses of cognitive impairment [2, 3].
Recently, deep learning models have demonstrated their capabilities in analyzing EHR data [4–8]. EHR data are a growing source of information that can be harnessed to provide an earlier diagnosis and to identify those at greatest risk for developing MCI. However, little is known about systematically analyzing patient data related to MCI in routinely-collected EHRs and how their temporal patterns are associated with the development of MCI. Although predicting MCI and understanding the progression from CU to MCI utilizing EHRs is a challenging and largely unexplored task, we believe deep learning models can be used to capture temporal characteristics of patient data in EHRs to predict early stages of MCI. In this study, we systematically investigated the application of EHR data analysis and deep learning models for predicting and clustering MCI patients.
II. Related works
Multiple studies to understand the progression from CU to MCI has been conducted. In [9], demographic, clinical, and neuropsychological measures implemented in Cox proportional hazard models were applied to predict progression from CU to MCI. The authors showed that MCI risk factors presented in their previous studies [10–13] can be used to predict MCI using multivariate models. They used the Mayo Clinic Study of Aging (MCSA) cohort [14, 15] and developed an augmented model capable of predicting progression from CU to MCI with AUC of 0.70. In [16], 224 CU participant were analyzed and followed up to detect measures or combination of measures that can be used to predict MCI. These researchers analyzed various MCI risk factors from different domains including cognitive, cerebrospinal fluid, magnetic resonance imaging (MRI), and genetic domain. They utilized time-dependent receiver operating characteristic and showed the feasibility of MCI prediction (best AUC using all variables > 0.83).
Another line of research is the prediction of dementia and understanding progression from MCI to dementia. Biomarkers, genetics, brain imaging as well as demographic and various variables related to individual’s lifestyle have been used in the literature to predict progression from MCI to dementia with AUC ranging from 0.48 to 0.91 [17]. In [18], [19] and [20] clinical variables were analyzed using logistic regression to predict progression of MCI to Alzheimer’s disease (AD) dementia. Researchers also utilized primary care data to predict dementia [21]. In [22], they used general practice data to create risk prediction models for dementia with sensitivity of 0.58 and specificity of 0.98 in the year before diagnosis.
With the availability of public databases related to AD dementia, such as the North American Alzheimer’s Disease Neuroimaging Initiative (ADNI) [23] and the European’s AddNeuroMed Study [24], big data has been utilized in AD research the past couple of years. Most of this research focused on diagnosing AD dementia, identify those at greatest risk of MCI, and predicting progression from MCI to AD dementia. Much of this work has utilized the non-community-based ADNI dataset and traditional machine learning models including support vector machines (SVMs), logistic regression and random forest [25]. SVM and linear models have been used in [26] to discriminate patients with AD from MCI and CU patients in ADNI dataset. Moreover, SVMs have been utilized in separating patients with AD dementia or MCI from CU patients using MRIs [27, 28], and predicting progression from MCI to AD dementia using AddNeuroMed dataset [29]. MRI images from ADNI has also been used in [30]. These results show that convolutional neural network based deep learning models can detect AD progression. In [31], neuroimaging, machine learning and deep learning has been used to predict conversion from MCI to AD in ADNI. XGBoost [32] showed the best performance in predicting MCI to AD progression in [31]. A comprehensive review of applying big data prospective and machine learning models to advance AD research is provided in [25].
Multiple studies examined the progression from CU or MCI to dementia. However, only a few studies considered progression from CU to MCI or MCI prediction using patients EHR data. Often, MCI is not well recorded in EHR data because it is not a clinical diagnosis per se, and thus there are not enough datasets suitable to train MCI predictive models. In addition, discriminating CU patients from patients with MCI is a very challenging task as MCI is the stage between the expected cognitive decline of normal aging and the more serious decline of dementia [33]. In this study, we addressed the above-mentioned challenges and demonstrated a potential of a deep learning model, coupled with natural language processing, to extract MCI risk factors and signals from unstructured EHRs and to predict onset of MCI. In addition, we utilized machine learning and deep learning techniques to visualize and cluster patients using EHR data and described a mechanism for EHR-based clustering.
III. Materials and methods
In this section, we first describe the dataset to train and test our models. Then, we describe the deep learning methods used in this study. This study includes two main components: MCI prediction and patient clustering. In MCI prediction, we used a deep learning technique, long short term memory (LSTM) [34] architecture described in section III.C to predict onset of MCI. In patient clustering, we used the denoising autoencoder described in section III.D to better represent the patient data. The outputs of this denoising autoencoder were visualized and clustered using t-Stochastic Neighbor Embedding (t-SNE) [35] and K-means algorithms [36].
A. Data
We used data obtain from the Mayo Clinic Study on Aging (n=5,923; 1,376 MCI) [15]. The MCSA is a prospective population-based cohort study with comprehensive periodic cognitive assessment (at baseline and repeated every 15 months), initiated in 2004 to investigate the epidemiology of MCI and dementia. Eligible subjects from the Olmsted County, Minn., population, were randomly selected and evaluated comprehensively in person using the clinical dementia rating scale, a neurological evaluation and neuropsychological testing. A consensus committee used previously published criteria to diagnose the participants with normal cognition, MCI or dementia.
MCSA participants have follow-up visits every 15 months and have accumulated more than 23,000 visits to date. In the current analyses, we only included MCI patients who progressed from CU to MCI; i.e., we excluded the patients who were diagnosed with MCI in their first visit to make sure we are predicting initial MCI diagnosis. The MCSA cohort mainly encounters Mayo Clinic, Olmsted County Medical Center, and Mayo Clinic Health System for the regular healthcare. This study used clinical notes to automatically extract MCI risk factors and signals. To simplify the study, we used patients who have any notes at Mayo Clinic—i.e., excluded patients who do not have any clinical notes at Mayo Clinic during each visit interval. This reduced the size of cohort (n=3,265; 558 MCI). The patient data after being diagnosed with MCI in MCSA were disregarded as we are aiming to predict MCI.
Different types of data were used to predict MCI: demographic information, diseases/disorders, and neuropsychiatric symptoms, and activity of daily living (ADL). Table I contains a complete list of variables and their EHR sources we used to train our models.
TABLE I.
LIST OF VARIABLES AND THEIR SOURCES.
| Variable Category | Variable | Source |
|---|---|---|
| Demographics | Age, Sex, Education | MCSA |
| Diseases / disorders | Hypertension, Atrial fibrillation, Angina, Congestive heart failure, Coronary artery disease, Myocardial infarction, Coronary artery bypass graft, Diabetes | Clinical notes |
| Neuropsychiatric symptoms | Delusion, Hallucinations, Agitation, Depression, Anxiety, Euphoria, Apathy, Disinhibition, Irritability/lability, Motor behavior, Appetite/eating change | Clinical notes |
| ADL | Bathing, Dressing, Feeding, Housekeeping, Responsible for own medication, Transportation, Toileting, Transferring, preparing food | Patient provided information |
| Others | Slow gait, cognitive complaint, impaired judgment/orientation, memory concern, difficulty for concentrating, difficulty for finance | Clinical notes |
Total 783,090 clinical notes were used to extract diseases/disorders, neuropsychiatric symptoms, and other types of data (Table I). To extract variables from clinical notes, we used the MedTaggerIE module in MedTagger [37], which is the open-source clinical natural language pipeline developed by Mayo Clinic for pattern-based information extraction with a capability of assertion detection (i.e., negated, possible, hypothetical, associated with a patient). We only included non-negated variables associated with patients.
B. Patient Representation
We represented patients both in a temporal and static mode and used them to train a temporal model (LSTM recurrent neural network) and a static model (random forest) for MCI prediction. To incorporate temporality, we converted the data into a visit-time format X(V, T), where V is patients’ visits and T is the visit dates, each visit vi includes all variables for a given visit listed in Table I, and each date ti is the relevant visit date. All of the patients’ visits for a period of 5 years before their first diagnosis dates for MCI patients and the latest visit for CU patients were used. We used a 15-month sliding window for the past 5 years of history of visits to make the temporal pattern asynchronous. Within each window an element-wise operation was used to combine the visits within the window.
We also represented patients in a static mode to train the static model. Instead of sliding a 15-month wide window, we used a 5-years window to cover the entire visit history of patients. As a result, the longitudinal data was converted to a matrix Y(P, L), where P is the complete list of patients, each pi is a vector including variables described in Table I and li is clinical diagnosis of MCI or CU of the patient pi.
C. Long-Short Term Memory (LSTM) Models
The LSTM model is one of the most powerful recurrent neural networks (RNNs) in processing longitudinal data efficiently incorporating time-varying variables. Fig. 1 shows a full and unrolled structure of RNN in which circles represent the network layers and the solid lines represent the weighted connections [34]. In a LSTM model, nodes are replaced with a unit called memory cell as shown in Fig. 2 [38]. A memory cell includes the input gate, output gate, and forget gate. The input gate decides how to update the cell state using the new input and the output gate determines how to filter the output. The forget gate decides which information the LSTM is going to forget; it considers both it and ht and then, utilizing a sigmoid function to generate a matrix with elements between 0 and 1. The previous cell state will be element-wisely multiplied by the numbers generated by the forget gate to determine how much information the LSTM unit wants to keep.
Fig 1.
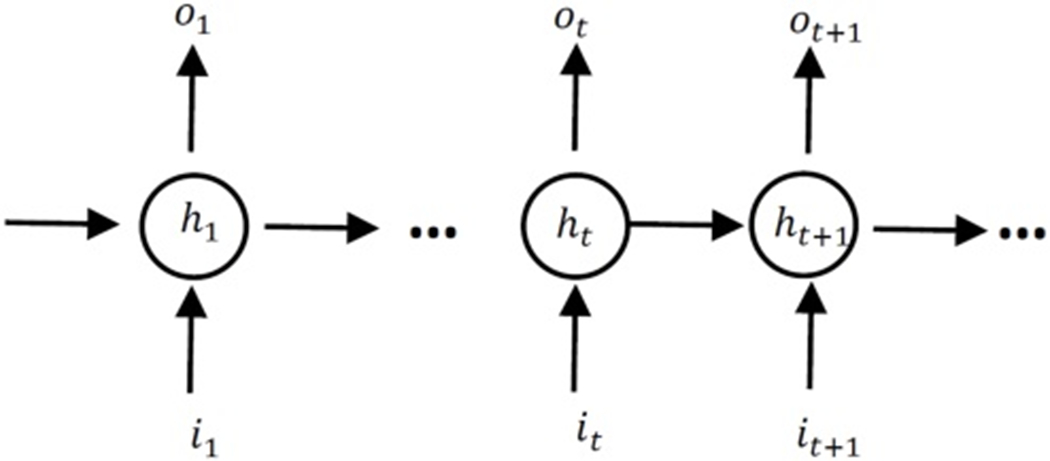
Unrolled structure of RNNs. The circles present hidden layers, it, ot and ht are respectively input, output and hidden state at time step t.
Fig 2.

A LSTM memory cell. Each LSTM unit includes the input gate, the output gate and the forget gate[39].
In this study, we utilized a dynamic LSTM to make the final predictions. We unroll the LSTM based on the number of time steps. The outputs of the last cell are then fed to a fully connected layer defined in (1).
| (1) |
Where, W is a n by m weight matrix in which n is the number of hidden neurons and m is the number of classes (two classes in this project), ot is the last output in the unrolled network, and b is a bias matrix. A threshold is used to make the final predictions based on the values of F. We optimized this threshold using a validation set during training.
D. Denoising Autuencoders
Denoising autoencoders [40] showed strong performance in efficiently representing participants data [41]. A four-layer denoising autoencoder was used in this study: an input layer, two hidden layers (encoder and decoder layers) and an output layer. Fig. 3 shows the architecture of the network used in this study. The first hidden layer encodes the input x using (2) and then the decoder decodes the encoded vector using (3).
Fig 3.

Artitecture of the denoising autoencoder to represent patients using HER
| (2) |
| (3) |
We corrupted 20 percent of the patient’s information and plugged the corrupted information as x in (2). The loss function is a mean squared error of the output layer and the patient’s information before the corruption.
Adam optimizer [42] at a learning rate of 0.01 was performed for 300 epochs to optimize the loss function. After training the autoencoder, all the samples were fed to the network without corruption and the outputs of the first hidden layer (y) were considered as a new representation for patient data. The new representation of data has lower dimension (we used 60 nodes for both of the hidden layers) and is less sparse. The dimensionality of trained autoencoder’s hidden nodes was further reduced using tSNE for visualization and clustering purpose. We used K-means clustering with k=5 as a default value because there are 5 potential clinical subtypes: 1) CU patients, 2) MCI positive- amnestic, single domain, 3) MCI positive- amnestic, multiple domain, 4) MCI positive- non amnestic, single domain, 5) MCI positive- non amnestic, multiple domain.
IV. Experimental results
A. Prediction of MCI
The LSTM models were built under the Tensorflow platform [43] and were trained using two Tesla K80 GPUs. For systematic training and testing, we used a randomized cross validation method to find optimal parameters. We first randomly split the data into training (70 percent of data), validation (10 percent) and testing sets (20 percent). We used a stratified approach to split the data into train, validation and test sets; the patients were split based on their age at the study entry to make sure that different age groups are equally represented across training, validation and testing datasets.
Each time we randomly selected a set of parameters in a predefined data pool; train a LSTM model on the training set and determine model parameters on the validation set. We repeated this process 100 times and selected the best parameter set on the validation set. Then, we used the best model (based on their performance on the validation set) to evaluate prediction performance on the unseen test data.
Learning rates were randomly selected from [10−6, 10−5, 10−4, 10−3, 10−2, 5−2, 8−-2, 10−1]. The number of hidden neurons were selected from a wide range of integers which includes [10, 20, 40, 60, 80, 100, 140, 180, 200, 300, 600]. The batch size was set to be 2n where n is an integer number in [5, 9]. The dropout probability was a random one decimal float number (%.1f) ranging from 0.3 to 0.7 and the number of iterations were selected from [104, 105, 106, 2 × 106, 5 × 106, 10 × 106, 15 × 106].
Random forests [44] serve as a baseline. Parameters of the random forests model were tuned using 5-fold cross validation. To search the hypothesis space, we used a random selection of parameters in a predefined pool of possible parameters. Number of estimators was a random integer number n × 100 where n is an even number in [2, 20]. We used the same unseen test data as used in LSTM model to evaluate the performance. It should be noted that random forest models are trained on the static data and LSTM models are trained on the longitudinal temporal data (refer to section III.B).
Performance of the LSTM and the random forest models are in Table II. All the results reported are on the test set. In Table II, RF and LSTM respectively indicate a random forest model and a LSTM model trained and tested on the original data. RF over-sampled and LSTM over-sampled are the same models but trained using an over-sampled training data and tested on the original test data; we increased the population of MCI patients using a sampling with replacement approach (i.e., we randomly selected MCI cases and added them to the training set to balance between MCI and CU cases).
TABLE II.
Performance of MCI prediction
| Model | Accuracy | Precision | Recall | F1 Score | AUC |
|---|---|---|---|---|---|
| RF | 0.82 | 0.44 | 0.13 | 0.21 | 0.73 |
| RF Over-sampled | 0.79 | 0.33 | 0.25 | 0.28 | 0.69 |
| LSTM | 0.73 | 0.33 | 0.59 | 0.43 | 0.71 |
| LSTM Over-sampled | 0.71 | 0.33 | 0.76 | 0.46 | 0.75 |
In Table II, the LSTM over-sampled produced both the highest F1-Score (0.46) and ROC AUC (0.75). Both LSTM and LSTM over-sampled performed better than the baselines in terms of F1 score. Both RF models produced higher accuracy than LSTM models but their recall were much lower than LSTM models, showing that random forest was biased due to the imbalanced class distribution (i.e., higher numbers of CU than MCI). These results indicate that LSTM models with temporal patterns of the data might have increased capability in predicting MCI compared to a traditional machine learning models using the static data.
In this study, we excluded the patients who were determined as MCI in their first visit to the MCSA. When including those MCI patients at the first visit (MCI=1,075), LSTM over-sampled produced higher performance than using the original data; i.e., precision, recall, F1-score and AUC were 0.53, 0.73, 0.61 and 0.74, respectively. This increased performance might be because there are more signals of patients who already had MCI before the first visit and/or the more data help create the more efficient deep learning models.
We also tested the random forests to investigate important risk factors for MCI. Random forests have a capability to identify the important variables based on impurity using information gain; i.e., how much each variable contributes to decreasing the impurity [44]. Table III shows the top 5 variables based on their effect in decreasing impurity.
TABLE III.
TOP 5 MCI RISK FACTORS DETERMINED BY RANDOM FORESTS
| Risk Factor | Score |
|---|---|
| Age | 0.16 |
| Hypertension | 0.08 |
| Education | 0.07 |
| Depression | 0.06 |
| Anxiety | 0.05 |
As can be seen from Table III, age is the most important feature in discriminating between MCI and CU patients. Age, hypertension, education, depression and anxiety have also been reported as important risk factors in developing MCI in other literature [21, 45–47].
B. Clustering and Visualizing MCI Patients
We utilized a denoising autoencoder (section III.D) with four layers to efficiently represent the patient data addressing data sparsity and high dimensionality. Reduced dimensionality enables the patient data to map a more meaningful space for better clustering. The outputs of the first hidden layer were mapped to a 2D space for visualization and clustering using tSNE and K-means. Fig. 4 visualizes the patient data. CU patients are indicated by blue dots and MCI patients are represented by red dots.
Fig 4.

Visualization of CU patients (blue dots) and patients with MCI (red dots).
Fig. 5 shows the distribution of MCI versus CU patients. Notably, ratio of MCI patients is much larger than the ratio of CU patients in cluster 2 but it is opposite in cluster 4. This may indicate the potential of a deep learning model for clustering distinct patient groups.
Fig 5.

Distribution of MCI versus CU patients within each cluster. Y-axis shows the ratio of CU or MCI population within the relevant cluster.
Fig. 6 (a) shows the distribution of males versus females and (b) further zoomed into males versus females for both CU and MCI patients. As can be seen, cluster 0 mostly includes males; almost 50 percent of males are clustered in this group. On the other hand, cluster 1 mostly includes females; half of the female population is clustered within this group.
Fig 6.
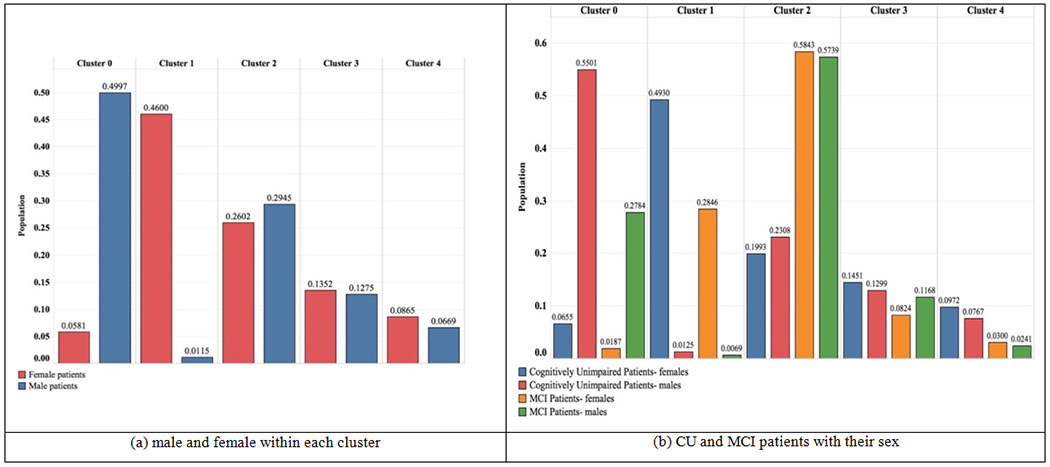
Distribution of male and female patients within each cluster.
V. Discussion
Despite the urgent need for early detection of MCI and dementia, there is a lack of automated tool available to healthcare providers using routine EHR data. Given that MCI is a precursor of dementia and could be a critical step in the prevention and control of AD and other dementias, it is crucial to find a more efficient way to determine MCI in its very early stages. This study addresses this issue by using routinely-collected EHR data as part of patient care for predicting onset of MCI.
The LSTM RNN demonstrated its potential for MCI prediction incorporating temporal patterns of patient data that were automatically extracted from EHRs. Although this study used limited set of MCI risk factors compared with the previous study that used manually annotated variables [9], our models still produced comparable (even slightly higher) performance. The LSTM RNN using longitudinal temporal data seems to be more efficient in predicting MCI compared to using the traditional machine learning models with static data. Machine learning techniques such as denoising autoencoders, K-means, and tSNE to visualize and cluster the patients EHR data also showed a good potential to identify certain patient groups. The predictive model and patient clustering could (have the potential to) assist in the clinic to support early identification of MCI patients as well as better characterization, determining more granular subgroups of MCI patients. This can enable tailored care plan and open up new clinical practice opportunities using routine EHR data.
The limitation of this study includes the use of only Mayo clinical notes even though participants enrolled in the MCSA visit other healthcare institutions, and thus the patients might not be ideally represented by their comprehensive longitudinal data. Use of EHR data from other institutions requires significant effort and this is aligned with our future work. We plan to use the medical record linkage system, the Rochester Epidemiology Project [48–50] to access other EHR data to compile comprehensive EHR data.
VI. CONCLUSION
A deep learning based model, LSTM RNN demonstrated a good potential incorporating temporal EHR patterns to predict the conversion from CU to MCI. When we combine this model with natural language processing to automatically extract MCI risk factors from EHR data, it could facilitate early detection of MCI addressing a current significant delay and thus improve treatment plans and health outcomes for patients.
The current model relies on a relatively small set of MCI patients and limited set of variables. In the future, we will use extended available and/or the entirety of patient EHR data instead of using known risk factors. We also plan to use publicly available data such as ADNI [23] for transfer learning or for testing the generalizability of the trained models. In addition, we will investigate multiple inputs multiple outputs models to enhance a prediction capability of MCI and dementia where the inputs are patient EHR data at different time steps and the outputs are MCI or dementia prediction at each time step.
Acknowledgment
This study was partially supported by NIAID R21AI 142702. The MCSA study was supported by NIH Grants U01 AG006786, P50 AG016574, the GHR Foundation, the Mayo Foundation for Medical Education and Research and was made possible by the Rochester Epidemiology Project (R01 AG034676).
CONFLICT OF INTERESTS
Maria Vassilaki receives research funding from NIH, Roche, and Biogen. Ronald C. Petersen consults for Roche, Inc, Merck, Inc, Biogen, Inc, Eisai, Inc; serves on the data and safety monitoring board for Genentech; and has given talks at national meetings for GE Healthcare.
Contributor Information
Sajjad Fouladvand, Department of Computer Science, Institute for Biomedical Informatics, University of Kentucky, Lexington, KY, USA.
Michelle M. Mielke, Division of Epidemiology, Department of Neurology, Mayo Clinic, Rochester, MN USA
Maria Vassilaki, Division of Epidemiology, Mayo Clinic, Rochester, MN USA.
Jennifer St. Sauver, Division of Epidemiology, Mayo Clinic, Rochester, MN USA
Ronald C. Petersen, Department of Neurology, Mayo Clinic, Rochester, MN USA
Sunghwan Sohn, Division of Digital Health Sciences, Mayo Clinic, Rochester, MN USA.
References
- [1].“Dementia statistics,” Alzheimer Disease International 2015, Available: http://www.alz.co.uk/research/statistics. [Google Scholar]
- [2].Goudarzvand S, Sauver JS, Mielke MM, Takahashi PY, and Sohn S, “Analyzing Early Signals of Older Adult Cognitive Impairment in Electronic Health Records,” in 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2018, pp. 1636–1640. [Google Scholar]
- [3].Goudarzvand S, Sauver J St., Mielke MM, Takahashi PY, Lee Y, and Sohn S, “Early temporal characteristics of elderly patient cognitive impairment in electronic health records,” BMC Medical Informatics and Decision Making, vol. 19, no. 4, p. 149, 2019/08/08 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Esteva A et al. , “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118, February 2 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cheng JZ et al. , “Computer-Aided Diagnosis with Deep Learning Architecture: Applications to Breast Lesions in US Images and Pulmonary Nodules in CT Scans,” Sci Rep, vol. 6, p. 24454, April 15 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rajkomar A et al. , “Scalable and accurate deep learning with electronic health records,” npj Digital Medicine, vol. 1, no. 1, p. 18, 2018/05/08 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shickel B, Tighe PJ, Bihorac A, and Rashidi P, “Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis,” IEEE J Biomed Health Inform, vol. 22, no. 5, pp. 1589–1604, September 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liang G, Fouladvand S, Zhang J, Brooks MA, Jacobs N, and Chen J, “GANai: Standardizing CT Images using Generative Adversarial Network with Alternative Improvement,” bioRxiv, p. 460188, 2018. [Google Scholar]
- [9].Pankratz VS et al. , “Predicting the risk of mild cognitive impairment in the Mayo Clinic Study of Aging,” Neurology, vol. 84, no. 14, pp. 1433–42, April 7 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mielke MM et al. , “Indicators of amyloid burden in a population-based study of cognitively normal elderly,” Neurology, vol. 79, no. 15, pp. 1570–7, October 9 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Mielke MM et al. , “Assessing the temporal relationship between cognition and gait: slow gait predicts cognitive decline in the Mayo Clinic Study of Aging,” J Gerontol A Biol Sci Med Sci, vol. 68, no. 8, pp. 929–37, August 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Geda YE et al. , “Prevalence of neuropsychiatric symptoms in mild cognitive impairment and normal cognitive aging: population-based study,” Arch Gen Psychiatry, vol. 65, no. 10, pp. 1193–8, October 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Roberts RO et al. , “Association of diabetes with amnestic and nonamnestic mild cognitive impairment,” Alzheimers Dement, vol. 10, no. 1, pp. 18–26, January 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Petersen RC et al. , “Prevalence of mild cognitive impairment is higher in men,” Neurology, vol. 75, pp. 889–897, 2010-09-07 00:00:00 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Roberts RO et al. , “The Mayo Clinic Study of Aging: design and sampling, participation, baseline measures and sample characteristics,” Neuroepidemiology, vol. 30, no. 1, pp. 58–69, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Albert M et al. , “Predicting progression from normal cognition to mild cognitive impairment for individuals at 5 years,” Brain, vol. 141, no. 3, pp. 877–887, March 1 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Stephan BC and Brayne C, “Risk factors and screening methods for detecting dementia: a narrative review,” J Alzheimers Dis, vol. 42 Suppl 4, pp. S329–38, 2014. [DOI] [PubMed] [Google Scholar]
- [18].Pozueta A et al. , “Detection of early Alzheimer’s disease in MCI patients by the combination of MMSE and an episodic memory test,” (in eng), BMC neurology, vol. 11, pp. 78–78, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gomar JJ, Conejero-Goldberg C, Davies P, Goldberg TE, and I. Alzheimer’s Disease Neuroimaging, “Extension and refinement of the predictive value of different classes of markers in ADNI: four-year follow-up data,” Alzheimers Dement, vol. 10, no. 6, pp. 704–12, November 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Poil SS, de Haan W, van der Flier WM, Mansvelder HD, Scheltens P, and Linkenkaer-Hansen K, “Integrative EEG biomarkers predict progression to Alzheimer’s disease at the MCI stage,” Front Aging Neurosci, vol. 5, p. 58, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ford E et al. , “Predicting dementia from primary care records: A systematic review and meta-analysis,” PLoS One, vol. 13, no. 3, p. e0194735, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ramakers IHGB et al. , “Symptoms of Preclinical Dementia in General Practice up to Five Years before Dementia Diagnosis,” Dementia and Geriatric Cognitive Disorders, vol. 24, no. 4, pp. 300–306, 2007. [DOI] [PubMed] [Google Scholar]
- [23].“Alzheimer’s Disease Neuroimaging Initiative,” ed, 2003.
- [24].Lovestone S et al. , “AddNeuroMed--the European collaboration for the discovery of novel biomarkers for Alzheimer’s disease,” Ann N Y Acad Sci, vol. 1180, pp. 36–46, October 2009. [DOI] [PubMed] [Google Scholar]
- [25].Zhang R, Simon G, and Yu F, “Advancing Alzheimer’s research: A review of big data promises,” (in eng), International journal of medical informatics, vol. 106, pp. 48–56, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gils M. v., Koikkalainen J, Mattila J, Herukka S, Lötjönen J, and Soininen H, “Discovery and use of efficient biomarkers for objective disease state assessment in Alzheimer’s disease,” in 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, 2010, pp. 2886–2889. [DOI] [PubMed] [Google Scholar]
- [27].Li M et al. , “An efficient approach for differentiating Alzheimer’s disease from normal elderly based on multicenter MRI using gray-level invariant features,” PLoS One, vol. 9, no. 8, p. e105563, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kohannim O et al. , “Boosting power for clinical trials using classifiers based on multiple biomarkers,” Neurobiol Aging, vol. 31, no. 8, pp. 1429–42, August 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lovestone S, Francis P, and Strandgaard K, “Biomarkers for disease modification trials--the innovative medicines initiative and AddNeuroMed,” J Nutr Health Aging, vol. 11, no. 4, pp. 359–61, Jul-Aug 2007. [PubMed] [Google Scholar]
- [30].Li R et al. , “Deep learning based imaging data completion for improved brain disease diagnosis,” Med Image Comput Comput Assist Interv, vol. 17, no. Pt 3, pp. 305–12, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shmulev Y and Belyaev M, “Predicting Conversion of Mild Cognitive Impairments to Alzheimer’s Disease and Exploring Impact of Neuroimaging,” in Graphs in Biomedical Image Analysis and Integrating Medical Imaging and Non-Imaging Modalities, Cham, 2018, pp. 83–91: Springer International Publishing. [Google Scholar]
- [32].Chen T, “XGBoost: A Scalable Tree Boosting System,” in 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, California, USA, 2016, pp. 785–794: ACM. [Google Scholar]
- [33].Petersen RC, “Mild cognitive impairment as a diagnostic entity,” Journal of Internal Medicine, vol. 256, no. 3, pp. 183–94, 2004. [DOI] [PubMed] [Google Scholar]
- [34].Graves A, “Generating Sequences With Recurrent Neural Networks,” ArXiv e-prints, vol. abs/1308.0850, 2013. [Google Scholar]
- [35].van der Maaten L and Hinton G, “Visualizing data using t-SNE,” vol. 9, pp. 2579–2605, 2008. [Google Scholar]
- [36].Lloyd S, “Least squares quantization in PCM,” IEEE Transactions on Information Theory, vol. 28, no. 2, pp. 129–137, 1982. [Google Scholar]
- [37].Liu H et al. , “An information extraction framework for cohort identification using electronic health records,” AMIA Jt Summits Transl Sci Proc, vol. 2013, pp. 149–53, 2013. [PMC free article] [PubMed] [Google Scholar]
- [38].Zaremba W, Sutskever I, and Vinyals O, “Recurrent Neural Network Regularization,” ArXiv e-prints, vol. 1409.2329, 2014. [Google Scholar]
- [39].Fouladvand S et al. , “Predicting substance use disorder using long-term attention deficit hyperactivity disorder medication records in Truven,” Health Informatics Journal, p. 1460458219844075, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Vincent P, Larochelle H, Bengio Y, and Manzagol P-A, “Extracting and composing robust features with denoising autoencoders,” presented at the Proceedings of the 25th international conference on Machine learning, Helsinki, Finland, 2008. [Google Scholar]
- [41].Beaulieu-Jones BK and Greene CS, “Semi-Supervised Learning of the Electronic Health Record for Phenotype Stratification,” bioRxiv, p. 039800, 2016. [DOI] [PubMed] [Google Scholar]
- [42].Kingma DP and Ba J, “Adam: A Method for Stochastic Optimization,” CoRR, vol. abs/1412.6980, 2014. [Google Scholar]
- [43].Mart et al. , “TensorFlow: a system for large-scale machine learning,” presented at the Proceedings of the 12th USENIX conference on Operating Systems Design and Implementation, Savannah, GA, USA, 2016. [Google Scholar]
- [44].Breiman L, “Random Forests,” Machine Learning, vol. 45, no. 1, pp. 5–32, 2001/10/01 2001. [Google Scholar]
- [45].Petersen RC et al. , “Predicting Progression to Mild Cognitive Impairment,” Annals of Neurology, vol. 85, no. 1, pp. 155–160, 2019/01/01 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gracia-García P et al. , “Depression and incident Alzheimer disease: the impact of disease severity,” (in eng), The American journal of geriatric psychiatry : official journal of the American Association for Geriatric Psychiatry, vol. 23, no. 2, pp. 119–129, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Roberts R and Knopman DS, “Classification and epidemiology of MCI,” (in eng), Clinics in geriatric medicine, vol. 29, no. 4, pp. 753–772, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rocca WA, Yawn BP, St Sauver JL, Grossardt BR, and Melton LJ 3rd, “History of the Rochester Epidemiology Project: half a century of medical records linkage in a US population,” Mayo Clin Proc, vol. 87, no. 12, pp. 1202–13, December 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].St Sauver JL et al. , “Data resource profile: the Rochester Epidemiology Project (REP) medical records-linkage system,” Int J Epidemiol, vol. 41, no. 6, pp. 1614–24, December 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sauver JLS, Grossardt BR, Yawn BP, III LJM, and Rocca WA, “Use of a Medical Records Linkage System to Enumerate a Dynamic Population Over Time: The Rochester Epidemiology Project,” American Journal of Epidemiology, vol. 173, no. 9, pp. 1059–68, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]


