Advertisement
Research ArticleInfectious diseaseInflammation
Open Access |  10.1172/JCI177992
10.1172/JCI177992
Neutrophil-mediated hypoxia drives pathogenic CD8+ T cell responses in cutaneous leishmaniasis
Erin A. Fowler,1 Camila Farias Amorim,2 Klauss Mostacada,1 Allison Yan,1 Laís Amorim Sacramento,2 Rae A. Stanco,1 Emily D.S. Hales,1 Aditi Varkey,1 Wenjing Zong,3 Gary D. Wu,3 Camila I. de Oliveira,4,5 Patrick L. Collins,1 and Fernanda O. Novais1
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Fowler, E. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Farias Amorim, C. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Mostacada, K. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Yan, A. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Amorim Sacramento, L. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Stanco, R. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Hales, E. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Varkey, A. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Zong, W. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Wu, G. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by de Oliveira, C. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Collins, P. in: JCI | PubMed | Google Scholar
1Department of Microbial Infection and Immunity, College of Medicine, The Ohio State University, Columbus, Ohio, USA.
2Department of Pathobiology, School of Veterinary Medicine and
3Division of Gastroenterology and Hepatology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
4Instituto Gonçalo Moniz, FIOCRUZ, Salvador, Brazil.
5Instituto Nacional de Ciência e Tecnologia em Doenças Tropicais, Salvador, Brazil.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
Find articles by Novais, F. in: JCI | PubMed | Google Scholar
Published June 4, 2024 - More info
J Clin Invest. 2024;134(14):e177992. https://doi.org/10.1172/JCI177992.
© 2024 Fowler et al. This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Received: November 27, 2023; Accepted: May 17, 2024
-
Abstract
Cutaneous leishmaniasis caused by Leishmania parasites exhibits a wide range of clinical manifestations. Although parasites influence disease severity, cytolytic CD8+ T cell responses mediate disease. Although these responses originate in the lymph node, we found that expression of the cytolytic effector molecule granzyme B was restricted to lesional CD8+ T cells in Leishmania-infected mice, suggesting that local cues within inflamed skin induced cytolytic function. Expression of Blimp-1 (Prdm1), a transcription factor necessary for cytolytic CD8+ T cell differentiation, was driven by hypoxia within the inflamed skin. Hypoxia was further enhanced by the recruitment of neutrophils that consumed oxygen to produce ROS and ultimately increased the hypoxic state and granzyme B expression in CD8+ T cells. Importantly, lesions from patients with cutaneous leishmaniasis exhibited hypoxia transcription signatures that correlated with the presence of neutrophils. Thus, targeting hypoxia-driven signals that support local differentiation of cytolytic CD8+ T cells may improve the prognosis for patients with cutaneous leishmaniasis, as well as for other inflammatory skin diseases in which cytolytic CD8+ T cells contribute to pathogenesis.
-
Introduction
Cutaneous leishmaniasis is caused by Leishmania parasites and exhibits a wide range of clinical manifestations, from self-healing lesions to chronic, debilitating infections (1). No vaccines exist for leishmaniasis, and antiparasitic drugs are often ineffective (2). Although the parasites influence disease severity, immune responses are often the major driver of disease. For example, CD8+ T cells in lesions are recruited to target parasite-infected cells and kill infected host cells through granule-mediated cytotoxicity. However, the destruction of infected cells does not kill Leishmania but leads to the release of parasites that metastasize to distant cutaneous sites (3). In addition, cell death induced by CD8+ T cells activates the NLRP3 inflammasome, IL-1β release, and chronic neutrophil recruitment and enhances inflammation (3–6). The importance of this pathway is supported by our previous analysis of lesions from patients in whom the expression of granzyme B (GZMB), perforin 1 (PRF1), and IL1B was associated with treatment failure (7). While murine and human studies underscore the pathogenic consequences of cytotoxicity, the microenvironmental cues and downstream mechanisms that drive cytolytic programs in lesional CD8+ T cells remain unknown.
In response to Leishmania infection, CD8+ T cells are activated and expand in draining lymph nodes (dLNs). The activated cells then travel to inflammatory sites, where they kill infected cells. Although infections also induce the differentiation of effector CD4+ T cells in secondary lymphoid organs, CD8+ T cells with a cytotoxic effector function are generally absent in LNs (8–11). A prior study implicated programmed cell death 1 (PD-1) ligation on dLN CD8+ T cells in suppressing their expression of GzmB, a process that is likely advantageous, since it would limit the killing of antigen-presenting cells (11). While such findings suggest that CD8+ T cell cytolytic effector function is only triggered once CD8+ T cells enter inflammatory sites (12, 13), the local cues that drive this response remain unclear. As the CD8+ T cell cytolytic program can have both positive and negative consequences for the host (14–33), understanding the mechanisms that drive cytotoxic CD8+ T cell programs remains an important goal for unraveling the pathogenesis of numerous diseases such as alopecia, vitiligo, bullous pemphigoid, Stevens-Johnson syndrome, and toxic epidermal necrolysis, in which cytolytic CD8+ T cells contribute to pathogenesis (15, 32, 34–37).
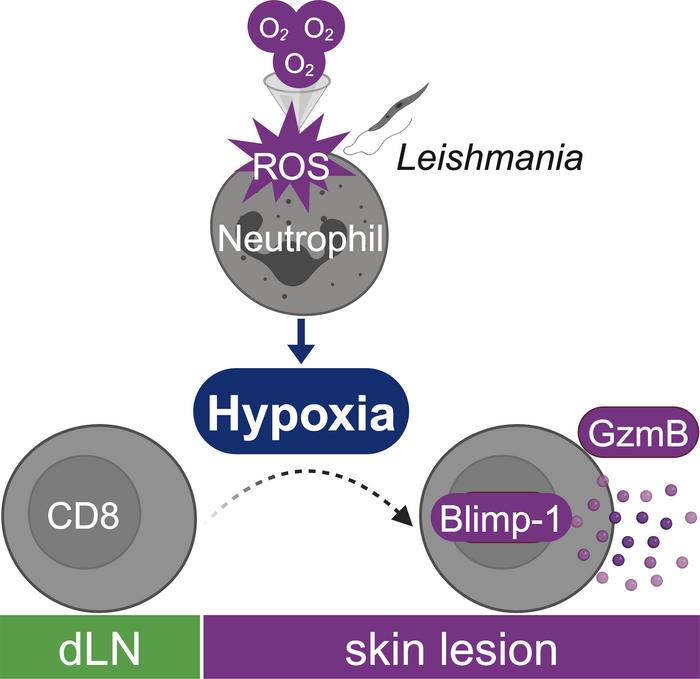
One potential mechanism relevant to inflamed tissues is hypoxia, which occurs when O2 consumption by infiltrating cells exceeds the supply (38) and differentially affects immune cell types. In Leishmania infection, hypoxia has been studied exclusively in macrophages and DCs (39, 40), where it can either promote or block effector responses, depending on the parasite species and the cell type (41–49). Here, we found that hypoxia within Leishmania-infected lesions directly promoted cytolytic effector function in newly recruited CD8+ T cells. Mechanistically, we identified a feed-forward loop, initiating with early recruitment of neutrophils that consume O2 to produce ROS, which promoted a persistently hypoxic environment, elevated the number of cytolytic CD8+ T cells, perpetuated neutrophil recruitment (4), and exacerbated tissue damage. Thus, our findings indicate that targeting cytolytic CD8+ T cells driven by hypoxic microenvironments will improve the prognosis for patients with cutaneous leishmaniasis and other inflammatory skin diseases.
-
Results
CD8+ T cells express GzmB in leishmanial lesions but not in the dLNs. Cytotoxic CD8 T cells are pathogenic in cutaneous leishmaniasis (3, 4, 6, 7, 50–54), but their anatomical distribution and mechanisms that regulate their induction remain unclear. In this regard, parasites are found in both dLNs and skin lesions. To determine whether CD8+ T cell cytotoxic effector function is present in both tissues, we assessed the expression of the cytotoxic effector molecule GzmB. C57BL/6 mice were infected with Leishmania in the ear, and 2 weeks after infection, the frequency of GzmBpos antigen-experienced (CD44hi) CD8+ T cells was assessed by flow cytometry. While GzmB-expressing CD8+ T cells were abundant in the infected skin, GzmB expression was nearly absent in the dLNs (Figure 1A). To test whether other effector functions by CD8+ T cells are intact in dLNs, we looked for IFN-γ production, which is required for protection against the parasite (1). As demonstrated previously (55), we detected IFN-γpos CD8+ T cells in dLNs, whereas IFN-γ expression was diminished in lesions (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI177992DS1). Collectively, these data demonstrate a tissue-specific effector response, with CD8+ T cells in dLNs exhibiting a protective phenotype through the production of IFN-γ and CD8+ T cells in lesions exhibiting a pathogenic phenotype by expressing GzmB. Since it is well described that cytotoxic CD8+ T cells are pathogenic in cutaneous leishmaniasis (3–7, 50, 51, 56, 57), we sought to determine how CD8+ T cells become cytotoxic. First, we asked if GzmBpos CD8+ T cells were preferentially recruited to the skin. To address this question, we treated mice with FTY720, a sphingosine 1-phosphate receptor agonist that blocks the egress of T cells from the dLNs. As expected, we observed a defect in the recruitment of T cells to the lesions of FTY720-treated mice compared with vehicle control–treated mice, as observed by a significant decrease in the frequency of CD3pos cells in lesions (Supplemental Figure 1B). Although FTY720-treated mice had fewer CD8+ T cells in lesions, those CD8+ T cells recruited to the inflamed skin still expressed GzmB (Figure 1B). If GzmBpos CD8+ T cells are preferentially recruited to cutaneous lesions, GzmBpos CD8+ T cells should accumulate in the dLNs of FTY720-treated mice. We found no differences in the frequency of GzmBpos CD8+ T cells in the dLNs of mice treated with FTY720 or vehicle (Figure 1B), suggesting that rather than preferential recruitment, exposure to the lesion microenvironment induced a cytotoxic profile in CD8+ T cells. To directly test this, we infected C57BL/6 CD45.1 or CD45.2 congenic mice with Leishmania and purified CD8+ T cells from dLNs of CD45.2 donor mice (GzmBneg) 3 weeks after infection. Donor dLN CD8+ T cells were then transferred directly into the lesions of infected CD45.1 animals (Figure 1C, schematic). Notably, we found that GzmBneg CD8+ T cells became GzmB-expressing CD8+ T cells in the lesions (Figure 1C). In contrast, CD8+ T cells that migrated back to dLNs remained GzmBneg (Figure 1C). These results demonstrated that the lesion microenvironment triggered cytotoxic programs in effector CD8+ T cells.
 Figure 1
Figure 1CD8+ T cell function is tissue specific. (A) GzmB expression in CD8+ T cells from dLNs and lesions from C57BL/6 mice in week 2 after infection with L. major. Data are representative of more than 3 experiments with at least 4 mice per experiment. (B) GzmB expression in CD8+ T cells from dLNs and lesions from L. major–infected C57BL/6 mice treated with FTY720 or vehicle daily for 10 days, 12 days after infection. Data are from 2 independent experiments with 5 mice per group. (C) Schematic representation of the transfer of purified CD8+ T cells from dLNs of CD45.2 mice into the lesions of CD45.1 recipients, both infected for 3 weeks. Flow cytometric plot shows GzmB expression in donor CD8+ T cells from dLNs and lesions from recipient mice 48 hours after transfer. Data were combined from 2 independent experiments. (D–H) RNA-Seq analyses of antigen-experienced (CD44hi) CD8+ T cells purified from dLNs and lesions of L. major–infected mice in week 5. (D) PCA showing PC1 and PC2. (E) Volcano plot highlighting the top 5 DEGs. The orange line indicates an adjusted P value of 0.05. (F) Heatmap showing the expression of genes encoding transcription factors (TFs) associated with effector-like T cell functions and the FC between lesional and dLN CD8+ T cells. (G) GSEA (Biocarta) showing pathways enriched in dLNs (green) and lesions (purple). NES, normalized enrichment score. (H) Heatmap of hypoxia-related pathways enriched in lesional and dLN CD8+ T cells. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001, by 2-tailed Student’s t test (A , C, and F) and 1-way ANOVA (B).
To determine the mechanisms by which CD8+ T cell cytotoxic programs are induced in lesions, we performed RNA-Seq analysis on purified antigen-experienced CD8+ T cells from dLNs and lesions of mice infected with Leishmania for 5 weeks. Principal component analysis (PCA) showed that almost half (principal component 1 [PC1], 47%) of the differences on the whole transcriptional profiles were associated with the organ from which the CD8+ T cells were derived (Figure 1D). Differentially expressed gene (DEG) analysis with thresholds of a FDR of less than 0.05 and a fold change (FC) of greater than 1.5 between tissues revealed overexpression of 118 genes in CD8+ T cells from lesions compared with dLNs and 26 DEGs overexpressed in dLNs compared with lesions (Figure 1E, Supplemental Table 1, and Supplemental Figure 1, C and D). Gzmb was the top DEG in CD8+ T cells from lesions compared with dLNs, with an FDR of 0.002 (Figure 1E and Supplemental Figure 1C). Several transcription factors play important roles in effector CD8+ T cell biology, including BATF, ID-2, IRF4, Stat4, T-bet, Zeb2, and Blimp-1 (encoded by Prdm1) (58–60), and we found that Id2, Irf4, and Prdm1 expression levels were significantly higher in lesions than in dLNs and that Prdm1 had the most significant FC between tissues (>100 FC higher in lesions) (Figure 1F). To further investigate the signals received by CD8+ T cells within lesions, we performed gene set enrichment analysis (GSEA) (Supplemental Table 2), which revealed an array of immune-related pathways, including a hypoxia-related signature (Figure 1G). Hypoxia is a common feature of many inflammatory disorders and can alter CD8+ T cell function (38, 61). To determine whether hypoxia is a critical signature of lesional CD8+ T cells, we assessed other pathway databases, as well as the Harris et al. hypoxia-specific pathway described in ref. 62, to see if there was enrichment for a hypoxia signature in lesional compared with dLN CD8+ T cells. We found enrichment for multiple hypoxia-related signatures in CD8+ T cells in lesions (Figure 1H and Supplemental Table 2). These data indicate that, after their exit from dLNs, CD8+ T cells recruited to cutaneous leishmaniasis lesions were exposed to a hypoxic environment, inducing the expression of key cytotoxicity effector mediators, including Prdm1.
Cutaneous leishmaniasis lesions are hypoxic and alter CD8+ T cell function. Normal skin is naturally low in O2 (63, 64), and since inflammatory environments are frequently hypoxic (65), we hypothesized that the lesions would be hypoxic. To evaluate hypoxia in Leishmania-infected lesions, we used pimonidazole, a 2-nitroimidazole reporter molecule that is reductively activated and forms covalent bonds with macromolecules in hypoxic cells. Two weeks after Leishmania infection, we assessed pimonidazole staining in infected and contralateral ears by confocal microscopy. As expected, we found pimonidazole staining within the epidermis, dermis, and hair follicles of naive mice (Figure 2, A and B, top images). Within lesions, we observed enlargement of the epidermis and dermis associated with robust pimonidazole staining (Figure 2, A and B, bottom images). Pimonidazole staining was absent from ulcerated areas, suggesting tissue heterogeneity in the hypoxic state of lesions. Representative images at lower (Figure 2A and Supplemental Figure 2A) and higher (Figure 2B and Supplemental Figure 2B) magnification show the differences in pimonidazole staining distribution between the naive and infected tissues.
 Figure 2
Figure 2Hypoxia induces GzmB and Prdm1 expression in CD8+ T cells. (A and B) Immunofluorescence staining and confocal microscopy of horizontal sections from lesions of C57BL/6 mice infected with L. major for 2 weeks. Mice received pimonidazole 1 hour before euthanasia. The top panels show contralateral ears (naive skin), and the bottom panels show lesions from infected ears. Pimonidazole is shown in green and nuclear DAPI staining in blue. Representative images are from 2 naive ears and 3 infected ears. Scale bars: 200 μm (A) and 100 μm (B). (C) Partial pressure of oxygen (pO2) in mmHg in dLNs and lesions of C57BL/6 mice infected with L. major for 2 weeks. Data were combined from 2 independent experiments. (D) GzmB expression in CD8+ T cells from dLNs of L. major–infected C57BL/6 mice; cells were cultured with DMOG or vehicle. (E) FC of Prdm1 mRNA over the average expression of vehicle-treated cells measured by qRT-PCR in DMOG- or vehicle-treated purified CD8+ T cells. (F) GzmB expression in CD8+ T cells from dLNs of C57BL/6 mice infected with L. major; cells were cultured under normoxia (21% O2) or hypoxia (1% O2). (G) FC of Prdm1 mRNA over the average expression of normoxic cells measured by qRT-PCR in purified CD8+ T cells cultured in normoxia or hypoxia. (D–G) Data shown are representative of more than 3 experiments with at least 4 mice per experiment. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001, by 2-tailed Student’s t test (C–G).
To compare O2 levels in lesions and dLNs, we infected mice with Leishmania, and 2 weeks after infection, mice received Oxyphor G4, a phosphorescent probe quenched by O2 (66). Infected skin showed a significant decrease in O2 compared with dLNs (Figure 2C). These data directly demonstrated that skin lesions present a hypoxic microenvironment to CD8+ T cells upon their recruitment from dLNs.
We tested whether exposure to hypoxia is sufficient to promote GzmB expression in dLN CD8+ T cells. We isolated dLN cells from infected mice and stimulated them with anti-CD3 and anti-CD28 antibodies in the presence of dimethyloxalylglycine (DMOG), a compound that mimics hypoxia at normal O2 tension. As shown in Figure 2D, DMOG induced the expression of GzmB in CD8+ T cells, further demonstrating that mimicking hypoxia promotes upregulation of their cytolytic program. Notably, DMOG treatment also increased Prdm1 mRNA expression compared with vehicle-treated CD8+ T cells (Figure 2E). We confirmed these findings by exposing CD8+ T cells to 1% O2 and found that hypoxia induced GzmB and Prdm1 expression in CD8+ T cells (Figure 2, F and G). We also tested whether hypoxia had a similar effect on naive and antigen-experienced CD8+ T cells (Supplemental Figure 3A) and we found that hypoxia enhanced GzmB expression only in antigen-experienced CD8+ T cells (Supplemental Figure 3B). These results raise the question of whether CD8+ T cells in vivo require Leishmania antigen to express GzmB or if sustained hypoxia alone is sufficient to promote GzmB. To test this, we infected mice with a T cell conditional deletion of Von Hippel–Lindau (referred to here as VHLcKO mice), which is necessary for the degradation of hypoxia-inducible factors (HIFs). Consequently, VHLcKO mice have HIF stabilization even in normoxic conditions. VHL deficiency did not increase GzmB expression in naive skin (Supplemental Figure 3C), suggesting that without Leishmania, there was no induction of GzmB when cells are forced to respond to hypoxia. In infected ears, VHL deletion increased GzmB expression in CD8+ T cells (Supplemental Figure 3C). One possible explanation is that at least a portion of CD8+ T cells in lesions were not exposed to hypoxia, which agrees with our observation that there were varied degrees of pimonidazole staining in the infected skin (Figure 2A). Another possibility is that CD8+ T cells newly recruited from the blood were not fully hypoxic and that forcing an immediate hypoxic response induced GzmB in these cells. Nevertheless, these data showed that antigen stimulation was necessary for CD8+ T cells to express GzmB in hypoxic conditions.
While the primary role of CD4+ T cells in our model is the production of IFN-γ and protection (1), CD4+ T cells can also be cytotoxic (67). Therefore, we tested whether hypoxia (1% O2) induces GzmB expression in CD4+ T cells from the dLNs of infected mice stimulated with anti-CD3 and anti-CD28 antibodies. Naive and antigen-experienced CD4+ T cells expressed more GzmB when exposed to hypoxia (Supplemental Figure 3D). However, GzmB expression was lower in CD4+ T cells than in CD8+ T cells (Supplemental Figure 3, B and D), suggesting that the induction of GzmB by hypoxia was shared by CD4+ and CD8+ T cells; however, the magnitude of the response was greater in CD8+ T cells. Given that CD4+ T cells were also exposed to the hypoxic microenvironment of lesions, we analyzed GzmB expression in CD4+ T cells in dLNs and lesions. We found that CD4+ and CD8+ T cells had a similar profile, with higher GzmB expression in lesions, although CD4+ T cells expressed significantly less GzmB than did CD8+ T cells (Supplemental Figure 3E). Collectively, these data demonstrate that the hypoxic state of lesions combined with antigen exposure stimulated the differentiation of recruited CD8+ T cells into cytotoxic effectors, with a moderate effect on the development of cytotoxic CD4+ T cells.
Blimp1 expression is restricted to GzmB-expressing CD8+ T cells. Blimp-1–deficient CD8+ T cells produce less GzmB (68), so we next sought to test cause-effect relationships between hypoxia, Blimp-1, and cytotoxicity. To this end, we infected Blimp-1 yellow fluorescent protein (YFP) reporter mice with Leishmania and assessed Blimp-1 expression in CD8+ T cells. We found that Blimp-1 was highly expressed in lesional CD8+ T cells but not in CD8+ T cells from the dLNs (Figure 3A). We also found that Blimp-1 expression was significantly higher in CD8+ T cells that expressed GzmB (Figure 3B), with similar results in CD4+ T cells (Supplemental Figure 4, A and B), suggesting a link between Blimp-1 and cytotoxicity in both T cell subsets. Since Blimp-1 regulates IL-10 production in CD4+ T cells (69), we measured the expression levels of IL-10 in dLNs and lesions of infected mice and found higher expression of IL-10 in lesional CD4+ and CD8+ T cells compared with dLNs (Supplemental Figure 4, C and D). In lesions, CD4+ and CD8+ T cells had higher Blimp-1 expression in IL-10pos cells than did their IL-10neg counterparts (Supplemental Figure 4, E and F), suggesting a strong relationship between Blimp-1 expression and IL-10, as previously described (69).
 Figure 3
Figure 3Blimp-1 expression is induced by hypoxia within skin lesions. (A) Expression of Blimp-1 in CD8+ T cells from dLNs and lesions from Blimp-1 YFP reporter mice infected with L. major for 1 week. Data from 2 independent experiments combined. (B) GzmB and Blimp-1 YFP expression in CD8+ T cells from lesions from WT or Blimp-1 reporter mice infected with L. major for 1 week. (C) Schematic representation of ODDCre Blimp-1fl/fl mice. (D) ODDCre Blimp-1fl/fl mice had been infected with L. major for 1 week when daily tamoxifen injections commenced. Mice were euthanized 2 weeks after infection. GzmB expression of lesional CD8+ T cells in tamoxifen-treated and untreated mice. Data were combined from 2 independent experiments. *P ≤ 0.05, **P ≤ 0.01, and ****P ≤ 0.0001, by 2-tailed Student’s t test (A, B, and D).
Finally, to assess whether GzmB expression in lesional CD8+ T cells resulted from hypoxic induction of Blimp-1, we used mice expressing a fusion protein bearing an oxygen-dependent degradation (ODD) domain from human HIF1A fused to a tamoxifen-inducible Cre recombinase gene (Cre/ERT2) (70). When expressed under normoxic conditions, the O2CreER fusion protein was degraded rapidly but was stabilized by hypoxia when combined with tamoxifen injection. These mice were crossed with Blimp-1fl/fl mice (referred to here as ODDCre Blimp-1fl/fl mice) to generate mice in which Blimp-1 expression is deleted specifically in hypoxic cells upon tamoxifen injection (Figure 3C). Since the skin has significantly less O2 than dLNs, these mice provide lesion-specific deletion of Blimp-1, thus ensuring the preservation of Blimp-1–dependent development of effector T cells in dLNs (normoxic) and their recruitment to the inflamed tissue. Leishmania-infected ODDCre Blimp-1fl/fl mice were treated with tamoxifen or left untreated for 1 week, and GzmB expression was evaluated. We found that the specific deletion of Blimp-1 in hypoxic cells significantly decreased GzmB expression in lesional CD8+ T cells compared with untreated mice (Figure 3D). Together, these results demonstrate that the upregulation of Blimp-1 by the hypoxic environment led to increased GzmB expression in CD8+ T cells.
A subset of GzmBpos CD8+ T cells express markers of exhaustion. Both hypoxia and Blimp-1 can induce the expression of many coinhibitory receptors involved in the exhaustion program (68, 71–74). To verify whether CD8+ T cells in lesions are exhausted, we reanalyzed a publicly available bulk RNA-Seq data set of 3 subsets of activated (stem-like CD101negTim3neg, transitory CD101negTim3pos and exhausted CD101posTim3pos) PD-1pos CD8+ T cells and naive CD8+ T cells in the context of chronic lymphocytic choriomeningitis virus (LCMV) infection. On the basis these subsets, we generated 3 specific gene signatures and compared them with our data set of CD8+ T cells isolated from the dLN and lesions (Figure 1, D–H). CD8+ T cells isolated from the lesions were enriched for all 3 signatures compared with CD8+ T cells isolated from the dLN (Supplemental Figure 5A). A subset of genes associated with exhaustion was higher in cells from lesions compared with dLNs, such as Pdcd1 (encoding for PD-1), Lag3 (encoding for Lag3), and Havcr3 (encoding for Tim3), while other genes — Tox, Cx3cr1, or Tcf7 — were not differentially represented in either tissue (Supplemental Figure 5B). We confirmed increased protein expression of PD-1, Lag3, and Tim3 in CD8+ T cells from lesions 2 weeks after infection (Supplemental Figure 5C). We sought to determine whether there was preferential expression of PD-1, Lag3, or Tim3 in CD8+ T cells that were GzmBpos and, therefore, had higher Blimp-1 expression and found that only Tim3 was preferentially expressed in GzmB-expressing CD8+ T cells (Supplemental Figure 5D). Collectively, these data suggest that coinhibitory molecule expression was a feature of some CD8+ T cells present in lesions and that a small subset of GzmBpos CD8+ T cells were either terminally exhausted or on their way to becoming so.
Blimp-1 expression is necessary for CD8+ T cells to mediate disease. On the basis of our data, we predicted that Blimp-1 expression is required for cytotoxic CD8 T cell–mediated disease in leishmaniasis. Blimp-1 plays a vital role in CD4+ T cell subsets (69, 75); hence, total T cell deletion of Blimp-1 would complicate interpretations. Therefore, we used our well-characterized mouse model of chronic leishmaniasis, in which severe disease develops following Leishmania infection of RAG–/– mice reconstituted with CD8+ T cells (3, 4, 55, 76). In this model, pathology is dependent on the ability of CD8+ T cells to be cytotoxic (3). RAG–/– mice infected with Leishmania were reconstituted with WT or CD8+ T cells lacking Blimp-1 expression (referred to here as Blimp-1cKO mice) or control animals receiving no T cells. As described previously (3), RAG–/– mice reconstituted with WT CD8+ T cells developed severe pathology characterized by GzmB-expressing CD8+ T cells in lesions, whereas RAG–/– mice that received no cells showed no signs of pathology (Figure 4A). Importantly, RAG–/– mice reconstituted with Blimp-1cKO CD8+ T cells had minimal disease (Figure 4A). As expected, there were similar numbers of parasites (Figure 4B) and a similar frequency of CD8+ T cells (Figure 4C) in lesions of RAG–/– mice reconstituted with WT and Blimp-1cKO CD8+ T cells. Notably, GzmB expression was significantly diminished in cells lacking Blimp-1 (Figure 4D). Collectively, these data indicate that Blimp-1 expression was driven by the hypoxic microenvironment of the lesion, triggering GzmB expression and CD8+ T cell–driven pathology.
 Figure 4
Figure 4Blimp-1 expression is required for CD8+ T cell–mediated disease. RAG−/− mice were infected with L. braziliensis and reconstituted with purified CD8+ T cells from WT or Blimp-1cKO mice. (A) Ear thickness and (B) parasite numbers in lesions at 7 weeks of infection. (C) CD8+ T cell frequency and (D) GzmB expression by CD8+ T cells in lesions were assessed directly ex vivo by flow cytometry 7 weeks after infection. Data represent 3 individual experiments with 3–5 mice per group. *P ≤ 0.05 and **P ≤ 0.01, by 2-tailed Student’s t test (A, C, and D).
Neutrophils generate the hypoxic microenvironment of cutaneous leishmaniasis lesions. Hypoxia occurs in tissues when O2 supply does not meet demand, including scenarios characterized by defective tissue vascularization or increased demand by cells present within a tissue (38). Leishmania-infected lesions are highly vascularized (77), so we hypothesized that O2 consumption by inflammatory cells recruited to lesions promotes hypoxia. Neutrophils are the first cells recruited to the skin upon Leishmania infection (78), but their recruitment does not control L. major parasites. Indeed, their chronic presence is associated with worsening disease (3, 4, 79–83). Therefore, we tested whether neutrophil recruitment contributed to the hypoxic state of inflamed skin. Leishmania-infected mice were injected with pimonidazole 1 hour before euthanasia at multiple time points after infection: (a) soon after parasite challenge (2 and 5 hours), (b) when lesions started to develop and CD8+ T cells were present and expressed GzmB (2 weeks), and (c) when lesions began to heal (9 weeks). As expected (78), neutrophils were quickly recruited after infection and decreased in frequency when lesions resolved (Figure 5A). Surprisingly, neutrophil (CD11bposLy6Gpos) staining by pimonidazole was significantly lower compared with other myeloid cells (CD11bposLy6Gneg) in the skin at all time points analyzed (Figure 5B). To confirm these observations, lesions from pimonidazole-injected mice were stained for Ly6G, revealing intense neutrophil recruitment in ulcered regions in the epidermis and clusters of neutrophils within the skin dermis (Figure 5C and Supplemental Figure 6A). Notably, regions without neutrophils had more pimonidazole staining than did those containing neutrophil clusters. We also observed that pimonidazole staining in regions surrounding neutrophil clusters was more intense. A quantitative assessment of pixel intensities (Supplemental Figure 6B) revealed a negative correlation between pimonidazole and Ly6G expression (Figure 5D). Higher magnification of regions with intense neutrophil recruitment in 2 samples showed a lack of pimonidazole staining in neutrophils (Figure 5E and Supplemental Figure 6C).
 Figure 5
Figure 5Neutrophils consume oxygen from lesions and stimulate GzmB expression in CD8+ T cells. (A and B) C57BL/6 mice infected with L. major received pimonidazole 1 hour before euthanasia at 2 hours, 5 hours, 2 weeks, and 9 weeks. (A) Frequency of neutrophils in lesions and (B) pimonidazole expression in neutrophils (CD11b+Ly6Gpos) and other myeloid cells (CD11b+Ly6Gneg) in lesions. (C) Representative confocal microscopy image of Ly6G (pink) and pimonidazole (green) staining in skin infected for 2 weeks. Scale bar: 200 μm. (D) Pixel intensity of pimonidazole and Ly6G based on 27 regions (Supplemental Figure 6B). (E) Two representative confocal microscopy images of nuclear staining (DAPI, blue), Ly6G (pink), and pimonidazole (green) in lesions infected for 2 weeks. Scale bar: 100 μm. (F–I) C57BL/6 mice infected with L. major were injected every 3 days with anti-Ly6G (clone 1A8) antibody or isotype control until euthanasia 2 weeks after infection. (F) Frequency of neutrophils in lesions, (G) pimonidazole staining in CD8+ T cells in lesions, and (H) GzmB frequency and MFI in CD8+ T cells in lesions. (I) Lesion size and number of parasites in lesions. (J) Lesion size and number of parasites in lesions in WT and Cybb–/– mice infected with L. major for 2 weeks. (K) GzmB frequency and MFI in CD8+ T cells in the lesions of WT or Cybb–/– mice. *P ≤ 0.05, ***P ≤ 0.001, and ****P ≤ 0.0001, by 2-tailed Student’s t test (B, F, H, and K) and 1-tailed Student’s t test (G).
These results suggest that areas surrounding neutrophils were relatively hypoxic. Accordingly, we hypothesized that neutrophils are responsible for promoting the hypoxic state of the skin by competing for oxygen, thereby controlling CD8+ T cell effector function. To test this, we depleted mice of neutrophils for the first 2 weeks of infection using anti-Ly6G antibody (Figure 5F) and assessed pimonidazole and GzmB expression in CD8+ T cells. We found that neutrophil depletion decreased pimonidazole expression (Figure 5G) and the frequency and MFI of GzmB-expressing CD8+ T cells compared with isotype control–treated mice (Figure 5H), without changing lesion size or parasite numbers (Figure 5I). These data suggest that neutrophils consumed O2 in lesions, altering the phenotype of CD8+ T cells.
Activated neutrophils assemble NADPH oxidase to produce ROS, which requires O2 (84). Thus, we tested whether O2 consumption to generate ROS affected CD8+ T cell function. For this purpose, we compared the ability of lesional CD8+ T cells to express GzmB in mice sufficient (WT) or deficient in the gp91phox subunit of the NADPH oxidase (Cybb–/–). Although there was no change in lesion size or parasite numbers in the skin at week 2 after infection (Figure 5J), we found significantly lower frequencies of GzmB-expressing CD8+ T cells in Cybb–/– mice than in WT (Figure 5K), forging a link between NADPH oxidase–dependent ROS production and the development of pathogenic CD8+ T cells in cutaneous leishmaniasis lesions.
The magnitude of hypoxia correlates with the presence of neutrophils in human cutaneous leishmaniasis lesions. To determine whether hypoxia is a feature of human disease, we compared transcriptional signatures from bulk RNA-Seq analysis performed in whole blood and total skin from healthy individuals and patients with cutaneous leishmaniasis using previously published data sets (54, 85) (Figure 6A). To estimate the levels of hypoxia gene expression in clinical samples, we performed single-sample GSEA using the Harris et al. hypoxia gene signature (62) and referred to this variable as the “hypoxia score” (Supplemental Table 3). While there was no difference in the hypoxia scores between blood from patients with cutaneous leishmaniasis and healthy individuals (Figure 6B and Supplemental Table 4), the cutaneous leishmaniasis lesions were significantly more hypoxic than healthy skin (Figure 6C and Supplemental Table 3). Importantly, we observed that the degree of the hypoxia-related gene expression in patients’ lesions was variable. To understand the biological implication of this variability, we performed unsupervised hierarchical clustering (HC) to classify lesions and healthy skin samples according to their hypoxic gene expression (Figure 6D). HC analysis revealed 2 groups of lesion samples: (a) “baseline” (n = 22), with a hypoxia phenotype comparable to that of control skin samples, and (b) “hypoxic” (n = 12), with a high hypoxic phenotype relative to the entire cohort (Figure 6, C and D). The remaining lesions had an intermediate phenotype. PCA revealed that hypoxic lesions were robustly segregated from the baseline counterparts: permutational multivariate ANOVA (PERMANOVA) (Pr), Pr(> F) = 0.001) (Figure 6E). Gene Ontology performed for genes overexpressed in hypoxic versus baseline lesions (FDR > 0.01 and FC = 1.5 thresholds) revealed significant enrichment for neutrophil chemotaxis and signaling (Supplemental Tables 5 and 6). Indeed, estimation of cell abundances from this unstructured bulk RNA-Seq data revealed an increase in neutrophil counts in hypoxic versus baseline lesions (Figure 6F). The top DEGs between hypoxic compared with baseline lesions included genes associated with neutrophil chemotaxis and survival, such as CXCL5, CXCL8, CXCR1, CSF3, and genes encoding proinflammatory cytokines such as IL1B, IL6, and OSM (Figure 6G and Supplemental Table 5). Therefore, our findings in mice are supported by our analysis of patients’ samples, linking the presence of neutrophils to the degree of hypoxia in lesions.
 Figure 6
Figure 6The degree of hypoxia-related gene expression is associated with the presence of neutrophils in patients. (A) Schematics of data collection for the RNA-Seq analysis performed in whole blood and total skin punch biopsies in healthy individuals and patients with cutaneous leishmaniasis. (B) Hypoxia score calculated in blood from healthy individuals (HI) (n = 14) and patients with cutaneous leishmaniasis (CL) (n = 50) and in (C) human intact skin (n = 6) and cutaneous leishmaniasis lesions from patients (n = 51) based on the Harris et al. (62) hypoxia gene signature. (D) Unsupervised hierarchical clustering classification of human skin samples in “baseline” and “hypoxic” lesions according to their hypoxia gene phenotypes. (E) PCA of baseline and hypoxic cutaneous leishmaniasis lesions. (F) Neutrophil abundances estimated by the MCP-counter method. The Wilcoxon test was used for statistical analysis. (G) Scatter plot showing differential gene expression analysis between hypoxic versus baseline lesions (FDR > 0.01 and FC = 1.5). In blue are the top DGEs from this analysis. **P ≤ 0.01 and ***P ≤ 0.001, by 2-tailed Student’s t test (C and F). HI, healthy individuals.
-
Discussion
Cytolytic CD8+ T cells cause severe inflammation in various settings (14, 15, 21, 24, 31–34, 86), although the factors that promote cytolysis by CD8+ T cells in the tissue microenvironments are still poorly understood. Here, we show that neutrophils recruited to the skin upon Leishmania infection consumed O2 to produce ROS, generating chronic inflammatory hypoxia. The resultant competition for O2 changed the aerobic microenvironment and promoted the expression of Blimp-1 in recruited CD8+ T cells, which resulted in GzmB expression and tissue damage. Because numerous skin diseases are associated with cytotoxicity, our findings provide evidence that local inhibition of this pathway should be considered as a strategy to ameliorate these diseases.
Preferential expression of cytolytic genes by CD8+ T cells in peripheral tissues but not in dLNs is observed in numerous conditions. For example, in models of graft-versus-host disease and graft-versus-leukemia, GzmB is absent in dLNs but present in the skin (9, 11). Additionally, memory CD8+ T cells acquire cytolytic activity upon entry into nonlymphoid tissues in vesicular stomatitis viral infections (10). The site-specific function of CD8+ T cells is not limited to GzmB expression. For example, in mice chronically infected with Trypanosoma cruzi, CD8+ T cells in the spleen produced IFN-γ, whereas CD8+ T cells within the muscle did not (87). Similarly, as previously shown (55), we found that CD8+ T cells in dLNs produced IFN-γ, while CD8+ T cells in the skin produced very little IFN-γ. Since hypoxic DCs produce less IL-12 (46, 48), this could explain the decrease in CD8+ T cell IFN-γ production in lesions (55) and suggests that hypoxia may not only promote pathogenic responses but may also prevent protective ones. Conversely, in diseases in which cytotoxicity is protective and IFN-γ is pathogenic, hypoxia would be expected to have the opposite effect. Our confocal microscopy data suggest that hypoxia affected many other cells beyond CD8+ T cells. In support of this observation, we found that CD4+ T cells also expressed more GzmB when exposed to hypoxia; however, the relevance of these cytotoxic CD4+ T cells to cutaneous leishmaniasis disease is unknown. Therefore, it is likely that hypoxia has a much greater role in tissue immunity to Leishmania.
There is substantial clinical and experimental evidence that hypoxia contributes to tumor progression, metastasis, and high mortality (88). While low O2 levels indicate myeloid suppressor cell accumulation (89) and tumor cell metastasis (90), HIFs provide CD8+ T cells with greater antitumor potential (91–93). HIF increases CD8+ T cell effector function during chronic viral infections and reduces viral titers, leading to higher mortality (94). These findings agree with the observed increases in perforin expression and killing ability of CD8+ T cells cultured in hypoxic conditions (93, 95, 96). Similar results were observed in lupus skin disease, in which HIF drove GzmB expression but, in this case, was independent of hypoxia (97). Hypoxia also alters CD8+ T cell metabolism (95) and induces tissue residency (91, 98) and exhaustion (72–74) programs. One of the limitations of our work is that it does not address the effect of HIFs on GzmB expression, and it is possible that HIFs directly promote Gzmb. However, our data provide evidence of an indirect induction of GzmB by hypoxia through Blimp-1 expression. Consistent with our findings, hypoxia also triggered Blimp-1 expression in pancreatic tumor cells (90). Since Blimp-1 can instruct tissue residency and exhaustion markers (68, 99–101), and we found an increased exhaustion phenotype in CD8+ T cells in the skin, we speculate that hypoxia-driven Blimp-1 expression may also initiate the CD8+ T cell program of exhaustion within cutaneous leishmaniasis lesions, which requires further investigation.
The effect of hypoxia is contradictory, as it produces more cytotoxic CD8+ T cells, while suppressing the function of some myeloid cells (46, 48). This balance may also occur within the same cell: for example, hypoxic induction of Blimp-1 regulates the cytotoxic function in T cells and paradoxically increases the expression of the antiinflammatory cytokine IL-10. In some situations, Blimp-1–dependent IL-10 production by CD4+ T cells protects against tissue damage (69). In CD8+ T cells, the cytotoxic program induced by Blimp-1 appears to outweigh the antiinflammatory effect of IL-10 on cutaneous leishmaniasis.
Another immune cell type implicated in the hypoxia/cytotoxicity axis revealed in our study is neutrophils, which rapidly migrate to infection sites and, in many infections, eliminate invading microbes. However, it is well recognized that the effect of neutrophils extends beyond pathogen killing, as they also shape the function of other cells, consequently affecting disease outcomes. For example, neutrophils producing IL-10 (102) or expressing PD-L1 (103) suppress CD4+ T cells. Additionally, neutrophils suppress effector responses by tumor-specific T cells in LNs (104) and within tumors (105). In chronic viral infections, suppressive neutrophil subsets reduce the antiviral capabilities of CD8+ T cells (106). In contrast, neutrophils facilitate DC migration to dLNs in contact hypersensitivity and prime allergen-specific CD8+ T cells (107). In cutaneous leishmaniasis, neutrophils recruited to lesions can have conflicting functions depending on the parasite species and mouse model. For example, neutrophils kill Leishmania directly by extracellular trap formation or cooperate with macrophages to activate microbicidal activities (108, 109). In contrast, neutrophils reduce costimulatory molecule expression in DCs and silently deliver parasites to macrophages (78, 110, 111). Despite these varied effects, it is clear that chronic recruitment of neutrophils is associated with tissue destruction in cutaneous leishmaniasis (3, 81, 83, 112). Here, we uncovered a new role for neutrophils in cutaneous leishmaniasis lesions by showing that their O2 consumption caused a phenotypic switch in CD8+ T cells.
Cytotoxic CD8+ T cells promote increased inflammation by inducing excessive cell death in lesions and enhancing chronic neutrophil recruitment to the skin (3, 4). In what may be related findings, neutrophil depletion decreases the hypoxic state of herpes stromal keratitis lesions and ameliorates disease (113). In a mouse model of colitis, the respiratory burst by Gr1pos cells — encompassing neutrophils and inflammatory monocytes — induced hypoxia in intestinal epithelial cells, which was critical for disease resolution (114). One limitation of our work is that it does not address the effect of other innate cells and their ability to deplete lesions of oxygen, which could also affect CD8+ T cell function. However, our work strongly supports the idea that O2 consumption and neutrophil recruitment are modulators of CD8+ T cell GzmB expression, which fosters the chronic presence of neutrophils in lesions (4). While the negative effect of this cascade of events is clear in cutaneous leishmaniasis, it could be beneficial in other contexts.
To be successful, host-directed therapies for infectious diseases should dampen immune responses that damage tissues and must not interfere with pathogen control (2). Therefore, defining immune pathways that are pathogenic rather than protective is critical. In cutaneous leishmaniasis, cytotoxicity is an ideal therapeutic target, since it is associated with treatment failure in patients who receive antiparasitic drugs (7) and does not control Leishmania (3). Patients affected by cutaneous leishmaniasis are frequently treated with antiparasitic drugs that require daily injections, and because failure rates are high, many patients must undergo multiple rounds of treatment. Our data provide evidence that local inhibition of the cytolytic program can circumvent the use of systemic drugs, which cause considerable side effects, and that topical targets should be considered as a new therapeutic strategy for cutaneous leishmaniasis.
-
Methods
Sex as a biological variable. Our animal experiments were performed using male and female mice, and controls were sex matched, but sex was not specifically tested as a biological variable. Human donors of both sexes were used for RNA-Seq analysis, and no sex differences were detected.
Mice. C57BL/6 CD45.2 and CD45.1 mice were purchased from Charles River Laboratories, and Prdm1 YFP, ODDCre/ERT2 (70), dLckCre, Blimp-1fl/fl, VHLfl/fl, and RAG–/– (B6.12957-RAG1tm1Mom) mice were purchased from The Jackson Laboratory and crossed in our mouse facility. Cybb–/– mice (115) were obtained from Juhi Bagaitkar (Nationwide Children’s Hospital, Columbus, Ohio, USA). Experiments assessing Blimp-1 expression with the Blimp-1 YFP reporter were reproduced using Blimp-1 GFP reporter mice (a gift from Stephen Nutt, Walter and Eliza Hall Institute of Medical Research [WEHI], Melbourne, Australia). GzmbCre mice (obtained from John Wherry, University of Pennsylvania) have the human GZMB promoter directing Cre recombinase expression to activated T cells; therefore, it does not directly affect mouse GzmB expression. GzmbCre and Blimp-1fl/fl mice were crossed to generate the deletion of Blimp-1 in activated T cells, and here we called RAG-/- mice that recieved purified CD8+ T cells from GzmbCreBlimp-1fl/fl mice Blimp-1cKO. ODDCre/ERT2 and Blimp-1fl/fl mice were crossed to generate mice in which Blimp-1 deletion was restricted to hypoxic cells in tamoxifen-treated mice. dLckCre and VHLfl/fl mice were crossed to generate the deletion of VHL in T cells after positive selection in the thymus, and we refer to these mice as VHLcKO mice. dLckCre mice were used as WT controls. All mice were maintained in a specific pathogen–free environment at The Ohio State University or the University of Pennsylvania Animal Care Facilities.
Parasites. L. major (strain WHO/MHOM/IL/80/Friedlin) and L. braziliensis (MHOM/BR/01/BA788) were grown in Schneider’s insect medium (Gibco, Thermo Fisher Scientific) supplemented with 20% heat-inactivated FBS (MilliporeSigma) and 2 mM glutamine (Thermo Fisher Scientific). Metacyclic-enriched promastigotes were used for infection (116). Mice were infected with either 106 L. major or 105 L. braziliensis intradermally in the ear, and lesion progression was monitored weekly by measuring the diameter of the ear with a caliper.
Cell purification and adoptive transfer. For the intralesional cell transfer mouse model, CD45.2 and CD45.1 mice were infected with L. major. CD45.2 mice were euthanized 3 weeks after infection, and CD8+ T cells from dLNs were purified using a magnetic bead separation kit (Miltenyi Biotec). CD8+ T cells (106 cells) were injected into the lesions of CD45.1 mice. Two days after injection, the recipient mice were euthanized, and GzmB expression in CD8+ T cells in the ear and dLNs was analyzed by flow cytometry directly ex vivo. For the RAG–/– mouse model, splenocytes from WT or Blimp-1cKO mice were collected, RBCs were lysed with ACK lysing buffer (Lonza), and CD8+ T cells were purified using a magnetic bead separation kit (Miltenyi Biotec). CD8+ T cells (3 × 106 cells) were transferred intravenously into RAG–/– mice that were subsequently infected with L. braziliensis. Mice reconstituted with CD8+ T cells received 4 injections of 250 μg anti-CD4 (clone GK1.5, Bio X Cell, catalog BE0003-1) within the first 2 weeks.
In vivo treatment. FTY720 (Cayman Chemical) was administered intraperitoneally (3 mg/kg) daily for 10 days before euthanasia, starting at two days post-infection. To induce Cre expression in the ODDCre/ERT2 mice, tamoxifen (MilliporeSigma) was dissolved in corn oil and administered intraperitoneally (6.7 mg/kg) for 7 days following infection. Pimonidazole (Hypoxyprobe) was administered intraperitoneally (9 mg/mL, 200 μL) 1 hour before euthanasia. For neutrophil depletion, mice received every 3 days injections of 250 μg anti-Ly6G (clone 1A8, Bio X Cell, catalog BE0075-1) within the first 2 weeks.
Tissue single-cell suspension preparation. Infected ears were harvested, the dorsal and ventral layers of the ear were separated, and the ears were incubated in RPMI 1640 (Gibco, Thermo Fisher Scientific) with 250 μg/mL Liberase TL (Roche Diagnostics) for 60–90 minutes in a shaker at 37°C and 5% CO2. Ears were dissociated using a cell strainer (40 μm, BD Pharmingen), and an aliquot of the cell suspension was used for parasite titration. dLNs were homogenized using a cell strainer (40 μm, BD Pharmingen) to obtain single-cell suspensions.
Parasite titration. The parasite burden in the ears was quantified as described previously (117). Briefly, the homogenate was serially diluted and incubated at 26°C. The number of viable parasites was calculated from the highest dilution at which parasites were observed after 7 days.
In vitro stimulation. dLN single-cell suspensions from infected C57BL/6 mice were cultured with 5 μg/mL plate-bound anti-CD3 (clone 145-2C11, Invitrogen, Thermo Fisher Scientific) and 0.5 μg/mL soluble anti-CD28 (clone 37.51, Invitrogen, Thermo Fisher Scientific) at 37°C and 5% CO2 for 48 hours in RPMI 1640 containing 100 Units of penicillin and 0.1 mg/mL streptomycin (MilliporeSigma), 2 mM l-glutamine (Thermo Fisher Scientific), and 10% FBS (MilliporeSigma). Cells were cultured in 1 mM DMOG (MilliporeSigma) dissolved in DMSO or an equivalent volume of DMSO (vehicle) for 48 hours. For mRNA isolation, dLN single-cell suspensions from infected C57BL/6 mice were obtained, and CD8+ T cells were purified using a magnetic bead separation kit (Miltenyi Biotec) and cultured as described above. For in vitro hypoxia experiments, cells were subjected to either normoxic conditions (standard incubator) or 1% oxygen (Baker Ruskinn InvivO2 400) in an incubator equipped to replace oxygen with nitrogen.
Flow cytometric analysis. Before surface and intracellular staining, cell suspensions were stained with a LIVE/DEAD Fixable Aqua Dead Cell Stain kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Analysis was performed using FlowJo Software (Tree Star), and gates were drawn on the basis of the fluorescence minus one (FMO) control. The following antibodies were used: CD45 (clone 30-F11, catalog 47-0451-82), GzmB (clone GB11, catalog GRB05 or GRB04), CD11b (clone M1/70, catalog 48-0112-82), IFN-γ (clone XMG1.2, catalog 25-7311-82), IL-10 (clone JES5-16E3, catalog 12-7101-82), Tim3 (clone RMT3-23, catalog 416-5870-82), and CD4 (clone RM4-5, catalog 17-0042-83 or 47-0042-82) (all from Invitrogen, Thermo Fisher Scientific); CD3 (clone 17A2, catalog 741319) and CD44 (clone IM7, catalog 751414) (both from BD Biosciences); CD45.2 (clone 104, catalog 109813), CD45.1 (clone A20, catalog 110705), CD90 (clone 53-2, catalog 140306), CD8β (clone YTS156.7.7, catalog 126610), Ly6G (clone 1A8, catalog 127614), Lag3 (clone C9B7W, catalog 125226), and PD-1 (clone 29F.1A12, catalog 135224) (all from BioLegend). The stained cells were run on a BD FACSymphony A3 or a FACSCanto II (BD Biosciences).
Quantification of Prdm1 mRNA by quantitative reverse transcription PCR. RNA extraction of in vitro–stimulated CD8+ T cells was performed using the Nulceospin RNA Mini kit (Macherery-Magel), and extracted RNA was then used to prepare cDNA using iScript Reverse Transcription Supermix for reverse transcription quantitative PCR (RT-qPCR) (Bio-Rad) on an ABS Proflex Thermal cycler (Thermo Fisher Scientific). qRT-PCR was carried out on a C1000 RT-PCR system using iTaq Universal SYBR Green Supermix (all from Bio-Rad) and primers targeting Prdm1 (forward, 5′-TTCTCTTGGAAAAACGTGTG G-3′ and reverse, 5′-GGAGCCGGAGCTAGACTTG-3′). The qRT-PCR results were normalized to Actb (forward, 5′-CGCTGTATTCCCCTCCATCG-3′ and reverse, 5′-CCAGTTGGTAACAATGCCATGT-3′). All reactions were carried out in duplicate, and data are represented as the FC over the average expression of normoxic or vehicle-treated cells.
Histology. Two weeks after infection, the mice were euthanized, and infected skin (lesions) and contralateral (naive skin) ears were collected and immediately immersed in 2% paraformaldehyde (Thermo Fisher Scientific) at room temperature. Samples were rinsed and immersed in PBS (Gibco, Thermo Fisher Scientific) for 2 hours, followed by cryoprotection in 30% sucrose. Samples were preserved in OCT (Thermo Fisher Scientific), and serial sections of 15 μm were obtained on a Tissue-Tek Cryo3 Flex cryostat (Sakura Finetek). Sections were collected onto Superfrost Plus microscope slides (Thermo Fisher Scientific), air-dried, and stored at –80°C.
Immunofluorescence, image acquisition, and analysis. Briefly, tissue sections were blocked and incubated with rabbit anti–pimonidazole antisera (Hypoxyprobe, catalog Pab2627), followed by incubation with rat anti–mouse Ly6G (clone 1A8, BioLegend, catalog 127604). Finally, sections were incubated separately with Alexa Fluor 568 goat anti-rabbit (Invitrogen, Thermo Fisher Scientific, catalog A11011) and Alexa Fluor 488 goat anti-rat (Invitrogen, Thermo Fisher Scientific, catalog A11008). The sections were counterstained with the nuclear probe DAPI (Biotechne-Tocris), covered with ProLong Gold Antifade Mountant (Invitrogen, Thermo Fisher Scientific) and kept protected from the light until imaging. Representative images were acquired with ×10 and ×20 objectives on an Olympus FV3000 confocal microscope (Olympus Corporation). The color of the different structures was digitally inverted to better match the data presented in this work. Using ImageJ software, version 1.53 (NIH), 27 regions of interest (ROIs) measuring 600 × 40 μm were created. From the low signal edge to the other, the pixel intensity values of each ROI were obtained.
Transcriptional profiling of purified mouse CD8+ T cells. C57BL/6 mice were infected with L. major, and 5 weeks after infection, the mice were euthanized; cells from the infected ear and dLN cells were purified and stained for flow cytometric analysis. Antigen-experienced CD8+ T cells were purified using the FACSAria cell sorter (gating strategy: live, singlets, CD90+, CD8β+CD44hi), and RNA was extracted with the RNeasy Plus Mini Kit (QIAGEN) according to the manufacturer’s instructions and used to prepare Poly(A)+-enriched cDNA libraries with the Illumina TruSeq Stranded mRNA library prep workflow. Ribo-Zero Gold rRNA depletion (Illumina) removed the ribosomal content. Quality assessment and quantification of RNA preparations and libraries were performed using the Agilent 4200 TapeStation and the Qubit 3, respectively. Samples were sequenced on an Illumina NextSeq 500 to produce 75 bp single-end reads. The raw reads were mapped to the mouse reference transcriptome (Ensembl; Mus musculus version 108) using Kallisto, version 0.46.0, and MultiQC, version 1.9, was used to check the quality of the alignment.
All subsequent analyses were conducted using the statistical computing environment R, version 4.1.0, RStudio, version 1.4.1717, and Bioconductor, version 3.13. Normalized gene expression matrix available for download as a text file in the NCBI Gene Expression Omnibus (GEO) database (GSE193005). Transcript quantification data were summarized to genes using the BiomaRt and tximport package and normalized using the trimmed mean of M-values (TMM) method in edgeR. Genes with less than 1 counts per million (cpm) in at least 3 samples were filtered out. Normalized filtered data were variance stabilized using the voom function in limma, and DEG analysis was performed with linear modeling using limma after correcting for multiple testing with Benjamini-Hochberg FDR correction. The list of 7 genes encoding for transcription factors associated with effector-like T cell functions was gathered from (58–60). Hypoxic enrichment per RNA-Seq sample was calculated with single-sample GSEA using the GSVA R package. The 5 hypoxia-related signatures used to estimate those levels were downloaded from Harris et al. 2002 (62), Hallmark systematic name (sn) no. M5891, Biocarta sn no. M13324, Biocarta sn no. M14863, and Reactome sn no. M641. The gene expression signatures of “stem-like” (CD101negTim3neg), “transitory” (CD101negTim3pos), and “exhausted” (CD101posTim3pos) PD-1pos CD8+ T cells were built from the normalized counts data set published elsewhere (118). Here, the normalized counts data were filtered for genes expressing more than 1 cpm in at least 3 samples and normalized with the limma R package. Individual differential gene expression was performed between “stem-like” versus naive, “transitory” versus naive, and “exhausted” versus naive CD8+ T cells using the edgeR R package. The lists of the top 100 DEGs from each comparison were used to build the state-of-exhaustion signatures. Single-sample GSEA using the GSVA R package was used to calculate state-of-exhaustion enrichment scores of CD8+ T cells isolated from dLNs and lesions of L. major–infected mice.
Hypoxia-related transcriptional analyses on blood and lesion biopsies from healthy individuals and patients with cutaneous leishmaniasis. Data in Figure 6 are derived from published transcriptional profiling (54, 85). The analysis carried out for this current study was performed from the filtered, normalized gene expression matrix available for download as a text file in the NCBI GEO database (GSE162760 and GSE214397). All subsequent analyses were conducted similarly to the analysis used for the purified mouse CD8+ T cells RNA-Seq data set. Hypoxia scores for the human RNA-Seq data set were calculated using the Harris et al. gene signature (62). Unsupervised hierarchical clustering to classify lesion samples according to their “hypoxic scores” was performed with maximum distances and Ward’s D2 minimum variance. PERMANOVA statistical testing was performed with the vegan R package. GSEA was performed using Broad Institute software, version 4.3.2. Gene Ontology analyses were carried out using DAVID Bioinformatics Resources (2021 Update) from the National Institute of Allergy and Infectious Diseases (NIAID), NIH and biological process terms, clustering gene sets by annotation similarity. Microenvironment cell populations (MCP) counter and immundeconv R packages were combined to estimate neutrophil abundances from the unstructured RNA-Seq data set. Data visualization was mostly performed in GraphPad Prism, version 10 (GraphPad Software) and in R programming language using the tidyverse and gplots tools.
Statistics. For differences between 2 groups, statistical significance was determined using an unpaired 1- or 2-tailed Student’s t test or a paired Student’s t test. For multiple comparisons, 1-way ANOVA was performed. The specific transcriptional analysis section describes detailed statistical analysis for RNA-Seq. Differences were considered significant when P was less than or equal to 0.05. Data are presented as the mean ± SEM.
Study approval. This study was conducted according to the recommendations in the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). The IACUCs of The Ohio State University and the University of Pennsylvania approved the study protocol.
Data availability. The complete RNA-Seq data analysis, R code, and file inputs and outputs used to perform the transcriptional analysis presented in this work are available in the GitHub repository “Hypoxia_FON” (https://github.com/camilafarias112/Hypoxia_FON). For all data values for all graphs, see the Supporting Data Values file.
-
Author contributions
EAF, CFA, RAS, EDSH, KM, LAS, WZ, CIDO, GDW, PLC, and FON designed the research studies. EAF, CFA, RAS, EDSH, KM, LAS, WZ, FON, AV, AY, and PLC conducted experiments. EAF, CFA, KAM, WZ, and FON analyzed and interpreted the data. EAF, CFA, and FON wrote the manuscript. FON supervised the study.
-
Acknowledgments
This work was supported by NIH grant R01AI162711 (to FON); the Host Defense and Microbial Biology Program from the Infectious Diseases Institute at The Ohio State University (to FON); the NIH training program T32 AI165391 “Interdisciplinary Program in Microbe-Host Biology” (to EAF); the NIH training program T32 AI118684 (to WZ); and the Host-Microbial Analytic and Repository Core at the Center for Molecular Studies in Digestive and Liver Diseases (P30DK050306, to GDW). The authors thank Juhi Bagaitkar (Nationwide Children’s Hospital, Columbus, Ohio, USA) for providing the Cybb–/– mice.
Address correspondence to: Fernanda O. Novais, The Ohio State University, College of Medicine, 720 BRT 460 W. 12th Avenue, Columbus, Ohio 43210, USA. Phone: 1.614.366.9272; Email: Fernanda.Novais@osumc.edu.
-
Footnotes
WZ’s present address is: Division of Gastroenterology/Hepatology/Nutrition, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2024, Fowler et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2024;134(14):e177992.https://doi.org/10.1172/JCI177992.
-
References
- Scott P, Novais FO. Cutaneous leishmaniasis: immune responses in protection and pathogenesis. Nat Rev Immunol. 2016;16(9):581–592.
- Novais FO, et al. Host-directed therapies for cutaneous leishmaniasis. Front Immunol. 2021;12:660183.
- Novais FO, et al. Cytotoxic T cells mediate pathology and metastasis in cutaneous leishmaniasis. PLoS Pathog. 2013;9(7):e1003504.
- Novais FO, et al. CD8+ T cell cytotoxicity mediates pathology in the skin by inflammasome activation and IL-1β production. PLoS Pathog. 2017;13(2):e1006196.
- Novais FO, Scott P. CD8+ T cells in cutaneous leishmaniasis: the good, the bad, and the ugly. Semin Immunopathol. 2015;37(3):251–259.
- Crosby EJ, et al. Lymphocytic choriomeningitis virus expands a population of NKG2D+CD8+ T cells that exacerbates disease in mice coinfected with leishmania major. J Immunol. 2015;195(7):3301–3310.
- Amorim CF, et al. Variable gene expression and parasite load predict treatment outcome in cutaneous leishmaniasis. Sci Transl Med. 2019;11(519):eaax4204.
- Silverio JC, et al. CD8+ T-cells expressing interferon gamma or perforin play antagonistic roles in heart injury in experimental Trypanosoma cruzi-elicited cardiomyopathy. PLoS Pathog. 2012;8(4):e1002645.
- Santos E Sousa P, et al. Peripheral tissues reprogram CD8+ T cells for pathogenicity during graft-versus-host disease. JCI Insight. 2018;3(5):e97011.
- Marzo AL, et al. Cutting edge: migration to nonlymphoid tissues results in functional conversion of central to effector memory CD8 T cells. J Immunol. 2007;179(1):36–40.
- Michonneau D, et al. The PD-1 axis enforces an anatomical segregation of CTL activity that creates tumor niches after allogeneic hematopoietic stem cell transplantation. Immunity. 2016;44(1):143–154.
- Weisberg SP, et al. Tissue-specific immunity for a changing world. Cell. 2021;184(6):1517–1529.
- Schenkel JM, Pauken KE. Localization, tissue biology and T cell state - implications for cancer immunotherapy. Nat Rev Immunol. 2023;23(12):807–823.
- Ammar M, et al. Involvement of granzyme B and granulysin in the cytotoxic response in lichen planus. J Cutan Pathol. 2008;35(7):630–634.
- Hiroyasu S, et al. Granzyme B inhibition reduces disease severity in autoimmune blistering diseases. Nat Commun. 2021;12(1):302.
- Kaminski L-C, et al. Cytotoxic T cell-derived granzyme b is increased in severe plasmodium falciparum malaria. Front Immunol. 2019;10:2917.
- Voskoboinik I, et al. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol. 2015;15(6):388–400.
- Walch M, et al. Cytotoxic cells kill intracellular bacteria through granulysin-mediated delivery of granzymes. Cell. 2014;157(6):1309–1323.
- Eeka P, Phanithi PB. Cytotoxic T Lymphocyte Granzyme-b mediates neuronal cell death during Plasmodium berghei ANKA induced experimental cerebral malaria. Neurosci Lett. 2018;664:58–65.
- Ji NN, et al. CTL-derived granzyme B participates in hippocampal neuronal apoptosis induced by cardiac arrest and resuscitation in rats. Front Neurol. 2019;10:1306.
- Gleave A, Granville DJ. Granzyme B in autoimmune skin disease. Biomolecules. 2023;13(2):388.
- Dotiwala F, et al. Killer lymphocytes use granulysin, perforin and granzymes to kill intracellular parasites. Nat Med. 2016;22(2):210–216.
- Metkar SS, et al. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity. 2008;29(5):720–733.
- Shen Y, et al. Topical small molecule granzyme B inhibitor improves remodeling in a murine model of impaired burn wound healing. Exp Mol Med. 2018;50(5):1–11.
- Park S, et al. Granzyme A-producing T helper cells are critical for acute graft-versus-host disease. JCI Insight. 2020;5(18):e124465.
- Parasa VR, et al. Enhanced CD8+ cytolytic T cell responses in the peripheral circulation of patients with sarcoidosis and non-Löfgren’s disease. Respir Med. 2018;138S:S38–S44.
- Junqueira C, et al. Cytotoxic CD8+ T cells recognize and kill Plasmodium vivax-infected reticulocytes. Nat Med. 2018;24(9):1330–1336.
- Balin SJ, et al. Human antimicrobial cytotoxic T lymphocytes, defined by NK receptors and antimicrobial proteins, kill intracellular bacteria. Sci Immunol. 2018;3(26):eaat7668.
- Trivedi P, et al. Perforin facilitates beta cell killing and regulates autoreactive CD8+ T-cell responses to antigen in mouse models of type 1 diabetes. Immunol Cell Biol. 2016;94(4):334–341.
- Vičić M, et al. Cytotoxic T lymphocytes as a potential brake of keratinocyte proliferation in psoriasis. Med Hypotheses. 2016;87:66–68.
- Patra V, et al. Accumulation of cytotoxic skin resident memory T cells and increased expression of IL-15 in lesional skin of polymorphic light eruption. Front Med (Lausanne). 2022;9:908047.
- Chung WH, et al. Granulysin is a key mediator for disseminated keratinocyte death in Stevens-Johnson syndrome and toxic epidermal necrolysis. Nat Med. 2008;14(12):1343–1350.
- Haque A, et al. Granzyme B expression by CD8+ T cells is required for the development of experimental cerebral malaria. J Immunol. 2011;186(11):6148–6156.
- Xing L, et al. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat Med. 2014;20(9):1043–1049.
- Zloza A, et al. Engagement of NK receptor NKG2D, but not 2B4, results in self-reactive CD8+ T cells and autoimmune vitiligo. Autoimmunity. 2011;44(8):599–606.
- Jacquemin C, et al. NKG2D defines a subset of skin effector memory CD8 T cells with proinflammatory functions in vitiligo. J Invest Dermatol. 2020;140(6):1143–1153.
- van den Boorn JG, et al. Autoimmune destruction of skin melanocytes by perilesional T cells from vitiligo patients. J Invest Dermatol. 2009;129(9):2220–2232.
- Taylor CT, Scholz CC. The effect of HIF on metabolism and immunity. Nat Rev Nephrol. 2022;18(9):573–587.
- Schatz V, et al. Hypoxia, hypoxia-inducible factor-1α, and innate antileishmanial immune responses. Front Immunol. 2018;9:216.
- Charpentier T, et al. Hypoxia inducible factor 1α: A critical factor for the immune response to pathogens and Leishmania. Cell Immunol. 2016;309:42–49.
- Mahnke A, et al. Hypoxia in Leishmania major skin lesions impairs the NO-dependent leishmanicidal activity of macrophages. J Invest Dermatol. 2014;134(9):2339–2346.
- Bowlin A, et al. Hypoxia inducible factor signaling in macrophages promotes lymphangiogenesis in Leishmania major infection. Infect Immun. 2021;89(8):e0012421.
- Degrossoli A, et al. Hypoxia modulates expression of the 70-kD heat shock protein and reduces Leishmania infection in macrophages. J Biomed Sci. 2004;11(6):847–854.
- Colhone MC, et al. Effect of hypoxia on macrophage infection by Leishmania amazonensis. J Parasitol. 2004;90(3):510–515.
- Arrais-Silva WW, et al. Expression of hypoxia-inducible factor-1alpha in the cutaneous lesions of BALB/c mice infected with Leishmania amazonensis. Exp Mol Pathol. 2005;78(1):49–54.
- Hammami A, et al. HIF-1α hampers dendritic cell function and Th1 generation during chronic visceralleishmaniasis. Sci Rep. 2018;8(1):3500.
- Hammami A, et al. HIF-1α is a key regulator in potentiating suppressor activity and limiting the microbicidal capacity of MDSC-like cells during visceral leishmaniasis. PLoS Pathog. 2017;13(9):e1006616.
- Hammami A, et al. IRF-5-mediated inflammation limits CD8+ T cell expansion by inducing HIF-1α and impairing dendritic cell functions during leishmania infection. PLoS Pathog. 2015;11(6):e1004938.
- Mesquita I, et al. The absence of HIF-1α increases susceptibility to leishmania donovani infection via activation of BNIP3/mTOR/SREBP-1c axis. Cell Rep. 2020;30(12):4052–4064.
- Novais FO, et al. Granzyme B inhibition by tofacitinib blocks the pathology induced by CD8 T cells in cutaneous leishmaniasis. J Invest Dermatol. 2021;141(3):575–585.
- Novais FO, et al. Genomic profiling of human Leishmania braziliensis lesions identifies transcriptional modules associated with cutaneous immunopathology. J Invest Dermatol. 2015;135(1):94–101.
- Crosby EJ, et al. Engagement of NKG2D on bystander memory CD8 T cells promotes increased immunopathology following Leishmania major infection. PLoS Pathog. 2014;10(2):e1003970.
- Santos C da S, et al. CD8(+) granzyme B(+)-mediated tissue injury vs. CD4(+)IFNγ(+)-mediated parasite killing in human cutaneous leishmaniasis. J Invest Dermatol. 2013;133(6):1533–1540.
- Farias Amorim C, et al. Multiomic profiling of cutaneous leishmaniasis infections reveals microbiota-driven mechanisms underlying disease severity. Sci Transl Med. 2023;15(718):eadh1469.
- Novais FO, et al. CD8+ T cells lack local signals to produce IFN-γ in the skin during leishmania infection. J Immunol. 2018;200(5):1737–1745.
- Sacramento LA, et al. NKG2D promotes CD8 T cell-mediated cytotoxicity and is associated with treatment failure in human cutaneous leishmaniasis. PLoS Negl Trop Dis. 2023;17(8):e0011552.
- Amorim Sacramento L, et al. CCR5 promotes the migration of pathological CD8+ T cells to the leishmanial lesions. PLoS Pathog. 2024;20(5):e1012211.
- Chen Y, et al. Transcriptional and epigenetic regulation of effector and memory CD8 T cell differentiation. Front Immunol. 2018;9:2826.
- Chang JT, et al. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. 2014;15(12):1104–1115.
- Yu B, et al. Epigenetic landscapes reveal transcription factors that regulate CD8+ T cell differentiation. Nat Immunol. 2017;18(5):573–582.
- Lee P, et al. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat Rev Mol Cell Biol. 2020;21(5):268–283.
- Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47.
- Evans SM, et al. Oxygen levels in normal and previously irradiated human skin as assessed by EF5 binding. J Invest Dermatol. 2006;126(12):2596–2606.
- Evans NTS, Naylor PFD. The systemic oxygen supply to the surface of human skin. Respir Physiol. 1967;3(1):21–37.
- Colgan SP, et al. Hypoxia and innate immunity: keeping up with the HIFsters. Annu Rev Immunol. 2020;38:341–363.
- Friedman ES, et al. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc Natl Acad Sci U S A. 2018;115(16):4170–4175.
- Cenerenti M, et al. The era of cytotoxic CD4 T Cells. Front Immunol. 2022;13:867189.
- Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31(2):309–320.
- Montes de Oca M, et al. Blimp-1-dependent IL-10 production by Tr1 cells regulates TNF-mediated tissue pathology. PLoS Pathog. 2016;12(1):e1005398.
- Lin Q, et al. Generation of a hypoxia-sensing mouse model. Genesis. 2020;58(3-4):e23352.
- Zhu L, et al. Blimp-1 impairs T cell function via upregulation of TIGIT and PD-1 in patients with acute myeloid leukemia. J Hematol Oncol. 2017;10(1):124.
- Scharping NE, et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat Immunol. 2021;22(2):205–215.
- Bannoud N, et al. Hypoxia supports differentiation of terminally exhausted CD8 T cells. Front Immunol. 2021;12:660944.
- Ford BR, et al. Tumor microenvironmental signals reshape chromatin landscapes to limit the functional potential of exhausted T cells. Sci Immunol. 2022;7(74):eabj9123.
- Fu SH, et al. New insights into Blimp-1 in T lymphocytes: a divergent regulator of cell destiny and effector function. J Biomed Sci. 2017;24(1):49.
- Belkaid Y, et al. CD8+ T cells are required for primary immunity in C57BL/6 mice following low-dose, intradermal challenge with Leishmania major. J Immunol. 2002;168(8):3992–4000.
- Weinkopff T, et al. Leishmania infection induces macrophage vascular endothelial growth factor a production in an ARNT/HIF-dependent manner. Infect Immun. 2019;87(11):e00088-19.
- Peters NC, et al. In vivo imaging reveals an essential role for neutrophils in leishmaniasis transmitted by sand flies. Science. 2008;321(5891):970–974.
- Passelli K, et al. The impact of neutrophil recruitment to the skin on the pathology induced by leishmania infection. Front Immunol. 2021;12:649348.
- Pedraza-Zamora CP, et al. Th17 cells and neutrophils: Close collaborators in chronic Leishmania mexicana infections leading to disease severity. Parasite Immunol. 2017;39(4)
- Charmoy M, et al. The Nlrp3 inflammasome, IL-1β, and neutrophil recruitment are required for susceptibility to a nonhealing strain of Leishmania major in C57BL/6 mice. Eur J Immunol. 2016;46(4):897–911.
- Gonçalves-de-Albuquerque S da C, et al. The equivocal role of Th17 cells and neutrophils on immunopathogenesis of leishmaniasis. Front Immunol. 2017;8:1437.
- Gimblet C, et al. Cutaneous leishmaniasis induces a transmissible dysbiotic skin microbiota that promotes skin inflammation. Cell Host Microbe. 2017;22(1):13–24.
- Moghadam ZM, et al. From flies to men: ROS and the NADPH oxidase in phagocytes. Front Cell Dev Biol. 2021;9:628991.
- Farias Amorim C, et al. Localized skin inflammation during cutaneous leishmaniasis drives a chronic, systemic IFN-γ signature. PLoS Negl Trop Dis. 2021;15(4):e0009321.
- Turner CT, et al. Granzyme B contributes to barrier dysfunction in oxazolone-induced skin inflammation through E-cadherin and FLG cleavage. J Invest Dermatol. 2021;141(1):36–47.
- Leavey JK, Tarleton RL. Cutting edge: dysfunctional CD8+ T cells reside in nonlymphoid tissues during chronic Trypanosoma cruzi infection. J Immunol. 2003;170(5):2264–2268.
- Chouaib S, et al. Hypoxic stress: obstacles and opportunities for innovative immunotherapy of cancer. Oncogene. 2017;36(4):439–445.
- Chiu DKC, et al. Hypoxia inducible factor HIF-1 promotes myeloid-derived suppressor cells accumulation through ENTPD2/CD39L1 in hepatocellular carcinoma. Nat Commun. 2017;8(1):517.
- Chiou S-H, et al. BLIMP1 induces transient metastatic heterogeneity in pancreatic cancer. Cancer Discov. 2017;7(10):1184–1199.
- Liikanen I, et al. Hypoxia-inducible factor activity promotes antitumor effector function and tissue residency by CD8+ T cells. J Clin Invest. 2021;131(7):e143729.
- Veliça P, et al. Modified hypoxia-inducible factor expression in CD8+ T cells increases antitumor efficacy. Cancer Immunol Res. 2021;9(4):401–414.
- Cunha PP, et al. Oxygen levels at the time of activation determine T cell persistence and immunotherapeutic efficacy. Elife. 2023;12:e84280.
- Doedens AL, et al. Hypoxia-inducible factors enhance the effector responses of CD8(+) T cells to persistent antigen. Nat Immunol. 2013;14(11):1173–1182.
- Finlay DK, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med. 2012;209(13):2441–2453.
- Gropper Y, et al. Culturing CTLs under hypoxic conditions enhances their cytolysis and improves their anti-tumor function. Cell Rep. 2017;20(11):2547–2555.
- Little AJ, et al. HIF-1 regulates pathogenic cytotoxic T cells in lupus skin disease. JCI Insight. 2023;8(16):e166076.
- Hasan F, et al. Hypoxia acts as an environmental cue for the human tissue-resident memory T cell differentiation program. JCI Insight. 2021;6(10):e138970.
- Behr FM, et al. Blimp-1 rather than hobit drives the formation of tissue-resident memory CD8+ T cells in the lungs. Front Immunol. 2019;10:400.
- Mackay LK, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 2016;352(6284):459–463.
- Kragten NAM, et al. Blimp-1 induces and Hobit maintains the cytotoxic mediator granzyme B in CD8 T cells. Eur J Immunol. 2018;48(10):1644–1662.
- Doz E, et al. Mycobacteria-infected dendritic cells attract neutrophils that produce IL-10 and specifically shut down Th17 CD4 T cells through their IL-10 receptor. J Immunol. 2013;191(7):3818–3826.
- Rambault M, et al. Neutrophils encompass a regulatory subset suppressing T cells in apparently healthy cattle and mice. Front Immunol. 2021;12:625244.
- Glodde N, et al. Reactive neutrophil responses dependent on the receptor tyrosine kinase c-MET limit cancer immunotherapy. Immunity. 2017;47(4):789–802.
- Coffelt SB, et al. IL-17-producing γδ T cells and neutrophils conspire to promote breast cancer metastasis. Nature. 2015;522(7556):345–348.
- Volberding PJ, et al. Suppressive neutrophils require PIM1 for metabolic fitness and survival during chronic viral infection. Cell Rep. 2021;35(8):109160.
- Weber FC, et al. Neutrophils are required for both the sensitization and elicitation phase of contact hypersensitivity. J Exp Med. 2015;212(1):15–22.
- Novais FO, et al. Neutrophils and macrophages cooperate in host resistance against Leishmania braziliensis infection. J Immunol. 2009;183(12):8088–8098.
- Guimarães-Costa AB, et al. Leishmania amazonensis promastigotes induce and are killed by neutrophil extracellular traps. Proc Natl Acad Sci U S A. 2009;106(16):6748–6753.
- Ribeiro-Gomes FL, et al. Efficient capture of infected neutrophils by dendritic cells in the skin inhibits the early anti-leishmania response. PLoS Pathog. 2012;8(2):e1002536.
- van Zandbergen G, et al. Cutting edge: neutrophil granulocyte serves as a vector for Leishmania entry into macrophages. J Immunol. 2004;173(11):6521–6525.
- Gonzalez-Lombana C, et al. IL-17 mediates immunopathology in the absence of IL-10 following Leishmania major infection. PLoS Pathog. 2013;9(3):e1003243.
- Rao P, Suvas S. Development of inflammatory hypoxia and prevalence of glycolytic metabolism in progressing herpes stromal keratitis lesions. J Immunol. 2019;202(2):514–526.
- Campbell EL, et al. Transmigrating neutrophils shape the mucosal microenvironment through localized oxygen depletion to influence resolution of inflammation. Immunity. 2014;40(1):66–77.
- Pollock JD, et al. Mouse model of X-linked chronic granulomatous disease, an inherited defect in phagocyte superoxide production. Nat Genet. 1995;9(2):202–209.
- Späth GF, Beverley SM. A lipophosphoglycan-independent method for isolation of infective Leishmania metacyclic promastigotes by density gradient centrifugation. Exp Parasitol. 2001;99(2):97–103.
- Uzonna JE, et al. Low dose Leishmania major promotes a transient T helper cell type 2 response that is down-regulated by interferon gamma-producing CD8+ T cells. J Exp Med. 2004;199(11):1559–1566.
- Hudson WH, et al. Proliferating transitory T cells with an effector-like transcriptional signature emerge from PD-1+ stem-like CD8+ T cells during chronic infection. Immunity. 2019;51(6):1043–1058.
-
Version history
- Version 1 (June 4, 2024): In-Press Preview
- Version 2 (July 15, 2024): Electronic publication



Copyright © 2024 American Society for Clinical Investigation
ISSN: 0021-9738 (print), 1558-8238 (online)