20170315 Cloud Accelerated Genomics - Tel Aviv / Phoenix
- 1. Cloud Accelerated Genomics Allen Day, PhD // Science Advocate @allenday // #genomics #ml #datascience
- 2. Table of Contents Section 1 Section 2 Section 3 Throughout Getting from Research to Application… Faster What are the bottlenecks for translating research into products? Emphasis on information processing. From CompBio Research to CompBio Engineering Getting results, more of them, and predictably improving Data Integration - Cutting Edge Use Cases What’s happening right now in industry and academia? How to use Google Cloud? I’ll introduce specific cloud services, along with examples of how they’ve been used successfully. Compute Engine, Kubernetes, Dataflow, Cloud ML, Genomics API
- 3. How to Understand? Linear B is a syllabic script that was used for writing Mycenaean Greek, the earliest attested form of Greek. The script predates the Greek alphabet by several centuries. The oldest Mycenaean writing dates to about 1450 BC.
- 4. Hypothetico-Deductive Method (Iterative) Organize Analyze, Interpret, and Plan Choose Data Acquire
- 5. Hypothetico-Deductive Method (Iterative) Organize Analyze, Interpret, and Plan Choose Data Acquire Situation: Not enough data. No means to get more. Dead Language. Outcome: Cannot understand. Also: Passive learning. No feedback.
- 6. DNA Sequencing Value Chain %Effort 0 100 Pre-NGS ~2000 Future ~2020 Now Sboner, et al, 2011. The real cost of sequencing: higher than you think! Secondary Analytics Analytics, Intepretation, Planning Experiment Design DNA Sequencing
- 7. Human Genetics Scenario Sboner, et al, 2011. The real cost of sequencing: higher than you think! Secondary Analytics Analytics, Intepretation, Planning Experiment Design %Effort 0 100 DNA Sequencing Situation: Unlimited Free DNA Result: Slow to understand. Pre-NGS ~2000 Future ~2020 Now
- 8. Q: Why Slow to Understand? A1: Data Processing Sboner, et al, 2011. The real cost of sequencing: higher than you think! Secondary Analytics Analytics, Intepretation, Planning Experiment Design %Effort 0 100 DNA Sequencing Situation: We still have an analysis bottleneck Result: Slow to understand. Pre-NGS ~2000 Future ~2020 Now
- 9. 00:20 - Connecting… 01:22 - Link Established
- 11. GOOGLE CONFIDENTIAL Google Cloud Platform lets you run your apps on the same system as Google
- 12. GOOGLE CONFIDENTIAL So you can focus on what matters to your science
- 13. Google confidential │ Do not distribute Google is good at handling massive volumes of data uploads per minute users search index query response time 300hrs 500M+ 100PB+ 0.25s
- 14. Google confidential │ Do not distribute Google can is good at handleing massive volumes of genomic data uploads per minute users search index query response time 300hrs 500M+ 100PB+ 0.25s ~6WGS >100x US PhDs ~1M WGS 0.25s
- 15. Google confidential │ Do not distributeGoogle confidential │ Do not distribute Google Genomics August 2015
- 16. Google confidential │ Do not distribute Google Genomics is more than infrastructure General-purpose cloud infrastructure Genomics-specific featuresGenomics API Virtual Machines & Storage Data Services & Tools
- 17. Google confidential │ Do not distribute BioQuery Analysis Engine Medical Records Genomics Devices Imaging Patient Reports Baseline Study Data Private Data Pharma Health Providers … Google’s vision to tackle complex health data Public Data
- 18. Google confidential │ Do not distribute BioQuery Analysis Engine Medical Records Genomics Devices Imaging Patient Reports Baseline Study Data Private Data Pharma Health Providers … Google’s vision to tackle complex health data Public Data
- 19. CONFIDENTIAL & PROPRIETARY 3.75 TERABYTES PER HUMAN 1.00 TB GENOME 2.00 TB EPIGENOME 0.70 TB TRANSCRIPTOME 0.06 TB METABOLOME 0.04 TB PROTEOME ~1 MB STANDARD LAB TESTS 5-YR LONGITUDINAL STUDY BASELINE STUDY: BIG DATA ANALYSIS Validate a pipeline to process complex phenotypic, biochemical, and genomic data ● Pilot Study (N=200) ○ Determine optimal biospecimen collection strategy for stable sampling and reproducible assays ○ Determine optimal assay methodology ○ Validate quality control methods ○ Validate device data against surrogate and primary endpoints ● Baseline Study (N=10,000+) ○ 6 cohorts from low to high risk for cardiovascular and cancer ○ Characterize human systems biology ○ Define normal values for a given parameter in heterogeneous states ○ Predict meaningful events ○ Validate wearable devices for human monitoring ○ Characterize transitions in disease state
- 20. Public Datasets Project https://cloud.google.com/bigquery/public-data/ A public dataset is any dataset that is stored in BigQuery and made available to the general public. This URL lists a special group of public datasets that Google BigQuery hosts for you to access and integrate into your applications. Google pays for the storage of these data sets and provides public access to the data via BigQuery. You pay only for the queries that you perform on the data (the first 1TB per month is free)
- 21. Confidential & ProprietaryGoogle Cloud Platform 21 Platinum Genomes 1000 Genomes Medical (Human) Population-scale Genome Projects 1000 Bulls 10K Dog Genomes Veterinary / Agriculture Open Cannabis Project Genome To Fields Panzea (1000 Maize) AgriculturePersonal Genome Project Human Microbiome Project NCBI GEO Human 100K Cancer Genome Atlas Many Other Interesting Datasets...
- 22. Google confidential │ Do not distribute PI / Biologist : variant calls for the 1,000 genomes
- 23. Google confidential │ Do not distribute Information: principal coordinates analysis (1000 genomes)
- 24. Google confidential │ Do not distribute Knowledge: populations cluster together
- 25. Bioinformatics scientist: BigQuery enables fast tertiary analysis
- 26. Google Cloud Platform Dataflow + BigQuery Used for Extract, Transform, Load (ETL), analytics, real-time computation and process orchestration. cloud.google.com/dataflow Dataflow Run SQL queries against multi-terabyte datasets in seconds. cloud.google.com/bigquery BigQuery
- 27. Google Cloud Platform Dataflow + BigQuery Used for Extract, Transform, Load (ETL), analytics, real-time computation and process orchestration. cloud.google.com/dataflow Dataflow Run SQL queries against multi-terabyte datasets in seconds. cloud.google.com/bigquery BigQuery
- 28. Google Cloud Platform Dataflow + BigQuery
- 29. Google confidential │ Do not distribute Example: GATK Analysis Pipeline Old way: install applications on host kernel libs app app app app Makefiles, CWL, WDL (on a virtual machine)
- 32. Google confidential │ Do not distribute Example: GATK Analysis Pipeline Old way: install applications on host kernel libs app app app app Makefiles, CWL, WDL (on a virtual machine)
- 33. Google confidential │ Do not distribute Example: GATK Analysis Pipeline ● Decouple process management from host configuration ● Portable across OS distros and clouds ● Consistent environment from development to production ● Immutable images New way: deploy containers Old way: install applications on host kernel libs app app app app libs app kernel libs app libs app libs app Makefiles, CWL, WDL (on a virtual machine) Dockerflow: Dataflow + Docker Benefits
- 34. Google confidential │ Do not distribute Use Case: Reproducible Science with Docker ● Objective: Build a mutation-detection pipeline ● Provided to competitors ○ Training data set ○ Evalutation data set ● Competitors submit pipelines as Docker images to DREAM Challenge host, Sage Bionetworks ● Submitted pipelines were used to process unseen data set ● Post-competition, Docker images made public ● Incidentally, Google won this competition with a deep-learning based variant caller called DeepVariant cloud.google.com/genomics/v1alpha2/deepvariant
- 35. Confidential & ProprietaryGoogle Cloud Platform 35 An idealized version of the hypothetico-deductive model of the scientific method is shown. Various potential threats to this model exist (indicated in red), including hypothesizing after the results are known (HARKing) and lack of data sharing. Together these undermine the robustness of results, and may impact on the ability of science to self-correct. Threats to reproducible science. http://www.nature.com/articles/s41562-016-0021
- 36. > java -jar target/dockerflow*dependencies.jar --project=YOUR_PROJECT --workflow-file=hello.yaml --workspace=gs://YOUR_BUCKET/YOUR_FOLDER --runner=DataflowPipelineRunner To run it: Variant Calls Your Variant Caller 36PubSub Queue Sequencer DNA Reads Genomics API Genomics API BigQuery Your Other Tool
- 37. GraphConnect SF 2015 / Graphs Are Feeding The World, Tim Williamson, Data Scientist, Monsanto https://www.youtube.com/watch?v=6KEvLURBenM
- 38. GraphConnect SF 2015 / Graphs Are Feeding The World, Tim Williamson, Data Scientist, Monsanto https://www.youtube.com/watch?v=6KEvLURBenM
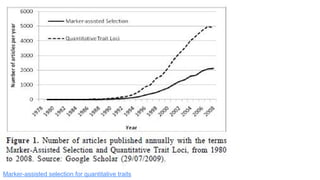
- 39. Marker-assisted selection for quantitative traits
- 40. Marker-assisted selection for quantitative traits https://www.sec.gov/Archives/edgar/data/1110783/0000950134 02011773/c71992exv99w2.htm
- 41. Google Cloud Platform Marker-Assisted Breeding Rapidly Increases Frequency of Favorable Genes https://www.slideshare.net/finance28/monsanto-082305a
- 42. Q: Why Slow to Understand? A1: Data Processing Sboner, et al, 2011. The real cost of sequencing: higher than you think! Secondary Analytics Analytics, Intepretation, Planning Experiment Design %Effort 0 100 DNA Sequencing Situation: We still have an analysis bottleneck Result: Slow to understand. Pre-NGS ~2000 Future ~2020 Now
- 43. Q: Why Slow to Understand? A2: Limited Feedback Sboner, et al, 2011. The real cost of sequencing: higher than you think! Secondary Analytics Analytics, Intepretation, Planning Experiment Design DNA Sequencing Situation: Data acquisition cost approaches zero However, still slow to understand, because: 1. Restricted choice of what can be observed, i.e. controlled modifications and artificial selection 2. Passive Learning. Limited feedback => Low rate of learning Contrast with active learning...
- 44. Act Observe Observe Act Orient Decide Decide Act Biological System Scientist Molecular Sensors: DNA sequencer, Mass spectrometer, Etc However... (Technology)-Limited Experimental Capability
- 45. Google Cloud Platform Even Moore’s Law / Carlson Curve
- 46. Google Cloud Platform Even Moore’s Law / Carlson Curve - also applies to writing DNA
- 47. Act Observe Observe Act Orient Decide Decide Act Biological System Scientist Molecular Sensors: DNA sequencer, Mass spectrometer, Etc Bioengineering Tech: DNA synthesizers, CRISPR/Cas9, Etc
- 48. Act Observe Observe Act Orient Decide Decide Act Biological System Scientist Molecular Sensors: DNA sequencer, Mass spectrometer, Etc Environmental Sensors: Laser scanners, Hyperspectral scanners, UAVs Etc Bioengineering Tech: DNA synthesizers, CRISPR/Cas9, Etc Regulate/Measure System I/O
- 49. Google Cloud Platform Integration with Geospatial, Management, and Terrestrial Sensor Data anezconsulting.com/precision-agronomy/
- 50. Google Cloud Platform Descartes Labs - Google Cloud Customer medium.com/@stevenpbrumby/corn-in-the-usa-d487dce84ee1 Cloud ML Engine TensorFlow
- 51. Google Cloud Platform Phenomobile, http://www.mdpi.com/2073-4395/4/3/349/htm See also: http://www.genomes2fields.org/
- 52. Google Cloud Platform Temporo-Spatial Imaging of Growing Plants
- 53. Google Cloud Platform Verily: Assisting Pathologists in Detecting Cancer with Deep Learning research.googleblog.com/2017/03/assisting-pathologists-in-detecting.html Prediction heatmaps produced by the algorithm had improved so much that the localization score (FROC) for the algorithm reached 89%, which significantly exceeded the score of 73% for a pathologist with no time constraint2 . We were not the only ones to see promising results, as other groups were getting scores as high as 81% with the same dataset. Model generalized very well, even to images that were acquired from a different hospital using different scanners. For full details, see our paper “Detecting Cancer Metastases on Gigapixel Pathology Images”.
- 54. 00:20 - Connecting… 01:22 - Link Established
- 55. Google Cloud Platform ~~)( , Cloud VisionTensorFlowGoogle Genomics Dataflow Cloud ML Engine Docker Baseline Study Data Private DataPublic Data
- 56. Build What’s Next Thank You! Allen Day, PhD // Science Advocate // @allenday // #genomics #ml #datascience