Translation
- 1. TRANSLATION M.Prasad Naidu MSc Medical Biochemistry, Ph.D,.
- 2. THE LANGUAGE OF NUCLEOTIDE SEQUENCE ON m-RNA ( GENTIC CODE ) IS TRANSLATED IN TO THE LANGUAGE OF AMINIO ACID SEQUENCE.
- 3. t-RNA ( soluble RNA) Transfer A.A from cytosol to the ribosomal protein synthesising machinery. 73 – 93 nucleotides in length
- 4. RIBOSOMES INFRASTRUCTURE FOR THE m-RNA, t- RNA & A.A TO INTERACT WITH EACH OTHER FOR TRANSLATION. 80S IN EUKARYOTES & 60S IN PROKARYOTES. LARGER 60S (50S) SUBUNIT SMALLER 40S (30S) SUBUNIT
- 5. Requirements for TRANSLATION: ribosomes m-RNA t-RNA A.As AMINOACYL t-RNA SYNTHETASES ATP, GTP ENZYMES OF TRANSLATION PROTEIN FACTORS
- 6. TRANSLATION: Steps 1. ACTIVATION OF A.A 2.INITIATION 3.ELONGATION 4.TERMINATION 5.POST TRANSLATIONAL PROCESSING
- 7. ACTIVATION OF A.A AMINOACYL t-RNA SYNTHETASE A.A + t-RNA + ATP Aminoacyl t-RNA + AMP
- 8. ACTIVATION OF A.A AMINOACYL t-RNA SYNTHETASE D ARM OF tRNA RECOGNISED BY THE ENZYME A.A + t-RNA + ATP Aminoacyl t-RNA + AMP ENZYME IDENTIFIES THE SPECIFIC t-RNA TRANSFERS AMINOACYL GROUP TO 3’ END CCA -COOH GROUP OF A.A ESTERIFIED WITH THE 3’ OH GROUP OF t-RNA. AT LEAST 1 ENZYME FOR 1 A.A
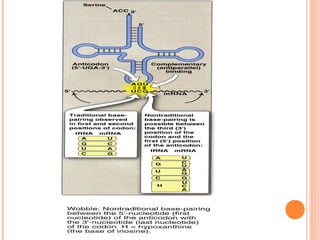
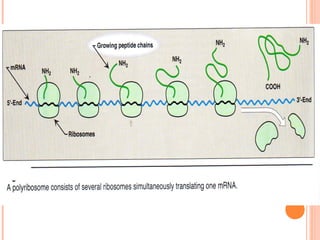
- 11. mRNA is read in 5’ to 3’ direction PROTEIN SYNTHESIS TAKES PLACE FROM N-TERMINAL TO C-TERMENAL END
- 12. INITIATION: Basic requirements for initiation Ribosome mRNA to be translated The initiating Met-tRNA A set of protein factors called initiation factors. In Eukaryotes there are at least nine factors, designated e If to indicate eukaryotic origin. In prokaryotes, three initiation factors are known IF1, IF2 and IF3
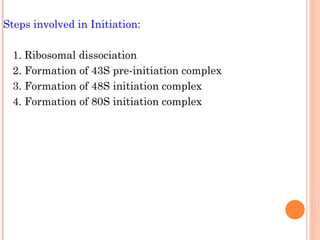
- 13. Steps involved in Initiation: 1. Ribosomal dissociation 2. Formation of 43S pre-initiation complex 3. Formation of 48S initiation complex 4. Formation of 80S initiation complex
- 14. Ribosomal dissociation: eIF3 and eIF1 A binds to 40S subunit and prevents the reassociation with 60S – causing ribosomal subunit dissociation.
- 15. 2. Formation of 43S pre-initiation complex GTP + eIF2 This complex binds to met-tRNA-I This GTP-eIF2-tRNA-I complex binds to the 40S ribosomal subunit to form 43S pre-initiation complex.
- 16. 3. Formation of 48S initiation complex Binding of m-RNA to 43S pre-initiation complex forms 48S initiation complex. 5’ end of m-RNA contains CAP which helps in binding of m- RNA to 43S pre-initiation complex. The association of mRNA with 43S initiation complex requires: CAP binding protein, eIF4, ATP KOZAK CONSENSUS (EUKARYOTES) on mRNA facilitates binding of mRNA to pre-initiation complex SHINE DALGARNO SEQUENCE on mRNA facilitates binding of mRNA to pre-initiation complex
- 17. 4. FORMATION OF 80S RIBOSOME ASSEMBLY 48S INITIATION COMPLEX + 60S RIBOSOME. + e-IF2 + e-IF5. e-IF 5 HAS GTPase ACTIVITY. GTP BOUND IN STEP 1 HYDROLYSED FOR ENERGY. ALL INITIATION FACTORS NOW RELEASED. NEXT STAGE IS ELONGATION.
- 18. 80S ribosome Ribosomal dissociation 40S subunit Ternary complex
- 19. 60S 43S Preinitiation complex 48S Inititation complex 80S Inititation complex Initiating codonCap eIF4 eIF eIF eIF
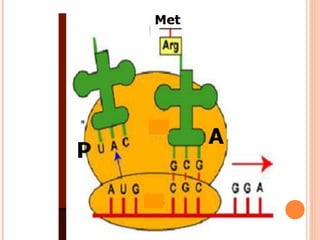
- 20. P & A SITES RIBOSOME CONTAINS 2 RECEPTOR SITES FOR t-RNA MOLECULES P (PEPTIDYL) SITE A (AMINOACYL) SITE Met-t-RNA OCCUPIES P SITE. ALL OTHER t-RNAs OCCUPY A SITE t-RNA BINDS WITH RIBOSOME THROUGH THE PSEUDOURIDINE ARM ANTICODON OF t-RNA & CODON ON m-RNA BIND A SITE IS FREE NOW.
- 24. ELONGATION: BINDING OF NEW AMINOACYL t-RNA CODON IN m-RNA DETERMINES THE INCOMING A.A EF 1 & GTP REQD. PEPTIDE BOND FORMATION: α-NH2 GROUP OF INCOMING A.A IN A SITE FORMS PEPTIDE BOND WITH COOH GROUP OF A.A IN P SITE. ENZYME IS PEPTIDYL TRANSFERASE NO NEED FOR ENERGY AS A.A IS ACTIVATED.
- 25. TRANSLOCATION: WHEN PEPTIDE BOND FORMED Met FROM P SITE SHIFTED TO A SITE. t RNA IS RELEASED FROM P SITE, NOW P SITE FREE RIBOSOME MOVES FORWARD BY 1 CODON PEPTIDYL t-RNA TRANSLOCATED TO P SITE.NOW A SITE EMPTY NEW AMINOACYL t-RNA WILL COME ONLY TO A SITE. STEP REQUIRES EF 2 AND ENERGY FROM GTP. NEW AMINOACYL t-RNA CAN COME IN ELONGATION CONTINUES….
- 26. APPP P P P AA A A A
- 27. ENERGY REQUIREMENTS: ACTIVATION OF A.A 2 ATP FOR PRE-INITIATION COMPLEX 1 GTP FOR INITIATION COMPLEX 1 ATP BINDING OF AMINOACYL t-RNA 1 GTP TANSLOCATION PROCESS 1 GTP
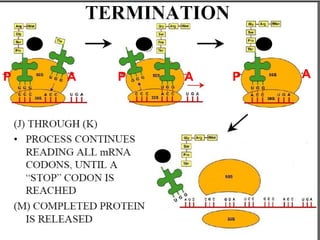
- 28. TERMINATION; SUCCESSIVE ADDITIONS OF A.A TERMINATION CODONS REACHED. NO t-RNA CORRESPONDING TO TERMINATION CODON A SITE REMAINS FREE RELEASING FACTOR ENTERS A SITE RELEASES THE PEPTIDE CHAIN WITH HYDROLYSIS OF GTP. RIBOSOME DISSOCIATES INTO 60 AND 40S SUBUNITS.
- 29. P P PA A A
- 32. PROTEIN TARGETTING: PROTEINS FOR EXTERNAL SECRETION: SYNTHESISED ON RER SIGNAL PEPTIDE (LEADER sequence) THRO ER, GOLGI REACHES CORRECT DESTINATION. INTERNAL PROTEINS: LACK SIGNAL PEPTIDE FREE RIBOSOMES DO NOT ENTER ER. ARE AVAILLABLE IN CYTOPLASM.
- 33. THANK YOU