Abstract
Free full text

Efficient discovery of overlapping communities in massive networks
Associated Data
Abstract
Detecting overlapping communities is essential to analyzing and exploring natural networks such as social networks, biological networks, and citation networks. However, most existing approaches do not scale to the size of networks that we regularly observe in the real world. In this paper, we develop a scalable approach to community detection that discovers overlapping communities in massive real-world networks. Our approach is based on a Bayesian model of networks that allows nodes to participate in multiple communities, and a corresponding algorithm that naturally interleaves subsampling from the network and updating an estimate of its communities. We demonstrate how we can discover the hidden community structure of several real-world networks, including 3.7 million US patents, 575,000 physics articles from the arXiv preprint server, and 875,000 connected Web pages from the Internet. Furthermore, we demonstrate on large simulated networks that our algorithm accurately discovers the true community structure. This paper opens the door to using sophisticated statistical models to analyze massive networks.
Community detection algorithms (1–17) analyze networks to find groups of densely connected nodes. These algorithms have become vital to data-driven methods for understanding and exploring network data such as social networks (4), citation networks (18), communication networks (19), and networks induced by scientific observation [e.g., gene regulation networks (20)].
Community detection is important for both exploring a network and predicting connections that are not yet observed. For example, by finding the communities in a large citation graph of scientific articles, we can make hypotheses about the fields and subfields that they contain. By finding communities in a large social network, we can more easily make predictions to individual members about who they might be friends with but are not yet connected to.
In this paper, we develop an algorithm that discovers communities in modern real-world networks. The challenge is that real-world networks are massive—they can contain hundreds of thousands or even millions of nodes. We will examine a network of scientific articles that contains 575,000 articles, a network of connected Web pages that contains 875,000 pages, and a network of US patents that contains 3,700,000 patents. Most approaches to community detection cannot handle data at this scale.
There are two fundamental difficulties to detecting communities in such networks. The first is that many existing community detection algorithms assume that each node belongs to a single community (1, 3–7, 14–16). In real-world networks, each node will likely belong to multiple communities and its connections will reflect these multiple memberships (2, 8–13, 17). For example, in a large social network, a member may be connected to coworkers, friends from school, and neighbors. We need algorithms that discover overlapping communities to capture the heterogeneity of each node’s connections.
The second difficulty is that existing algorithms are too slow. Many community detection algorithms iteratively analyze each pair of nodes, regardless of whether the nodes in the pair are connected in the network (5, 6, 10). Consequently, these algorithms run in time squared in the number of nodes, which makes analyzing massive networks computationally intractable. Other algorithms avoid computation about unconnected nodes (2–4, 7–9, 11–17). These methods are more efficient, but either make too simple assumptions, are still difficult to scale, or have difficulty with prediction.
Our algorithm addresses these difficulties. It discovers the hidden overlapping communities in massive networks, and its results can be used to explore, understand, and form predictions about their structure. Fig. 1 gives an example. This is a subgraph of a network of 575,000 scientific articles on the arXiv preprint server (21); each link denotes that an article cites or is cited by another article. Our algorithm analyzed this network, discovering overlapping communities among the citations. It assigned multiple communities to each article and a single community to each link. Many articles mostly link to other articles within their main community. However, the article “An alternative to compactification” (22) is different—it links to multiple communities, which suggests that it relates to multiple fields. Identifying nodes in large networks that bridge multiple communities is one way that our algorithm gives insights into the structure of the network.

The discovered community structure in a subgraph of the arXiv citation network (21). The figure shows the top four link communities that include citations to “An alternative to compactification” (22), an article that bridges several communities. We visualize the links between the articles and show some highly cited titles. Each community is labeled with its dominant subject area; nodes are sized by their bridgeness (39), an inferred measure of their impact on multiple communities. This is taken from an analysis of the full 575,000 node network.
Our algorithm identifies hundreds of overlapping communities among millions of nodes in a matter of hours. It is fast because of its simple structure: (1) subsample a subgraph from the full graph; (2) analyze the subgraph under the algorithm’s current estimate of the communities; (3) update this estimate of the communities, based on the analysis from the previous step; (4) repeat.
This powerful algorithmic structure is efficient because it only analyzes a subgraph of the network at each iteration. These subgraphs can be as large or as small as is computationally feasible, and can be designed to maximize the statistical information for efficiently finding communities. Furthermore, the algorithm does not require that the network be fully observed before beginning to estimate communities; its algorithmic structure naturally interleaves data collection with data analysis.
What we will show below is that our algorithm emerges when we take a Bayesian approach to detecting overlapping communities. In particular, we posit a probabilistic model of networks (23) where each node can belong to multiple communities (10). We then analyze a network by computing the posterior, the conditional distribution of the hidden communities given the observed network. The efficient structure of the algorithm—iteratively subsampling the network and updating an estimate of the hidden communities—emerges when we approximate this conditional distribution with variational methods (24) in combination with stochastic optimization (25, 26).
In the rest of the paper, we describe a model of overlapping communities (10) and present our efficient algorithm for computing with it. We demonstrate the capabilities of this analysis on three large real-world networks and report on a study of large simulated networks where the community structure is known.
One of the main advantages of taking a probabilistic approach to network analysis is that the models and algorithms are reusable in more complex settings. Our strategy for analyzing networks easily extends to other probabilistic models, such as those taking into account degree distribution or node attributes beyond the network. The approach we develop here opens the door to using sophisticated statistical models to analyze massive networks.
The Model and Algorithm
We describe a Bayesian model of overlapping communities and our scalable algorithm for computing with it.
A Mixed-Membership Stochastic Blockmodel.
We describe the model by its probabilistic generative process of a network. In this process, the community memberships will be encoded as hidden random variables. Given an observed network, such as a social network of friendship ties, we discover the hidden community structure by estimating its conditional distribution.
Classical community membership models, like the stochastic blockmodel (5, 6, 27), assume that each node belongs to just one community. Such models cannot capture that a particular node’s links might be explained by its membership in several overlapping groups, a property that is essential when analyzing real-world networks. Rather, our model is a type of “mixed-membership stochastic blockmodel” (10), a variant of the stochastic blockmodel where each node can exhibit multiple communities.
The model assumes there are K communities and that each node i is associated with a vector of community memberships  . This vector is a distribution over the communities—it is positive and sums to 1. For example, consider a social network and a member for whom one-half of her friends are from work and the other half are from her neighborhood. For this node,
. This vector is a distribution over the communities—it is positive and sums to 1. For example, consider a social network and a member for whom one-half of her friends are from work and the other half are from her neighborhood. For this node, 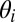 would place one-half of its mass on the work community and the other half on the neighborhood community.
would place one-half of its mass on the work community and the other half on the neighborhood community.
To generate a network, the model considers each pair of nodes. For each pair  , it chooses a community indicator
, it chooses a community indicator  from the ith node’s community memberships
from the ith node’s community memberships  and then chooses a community indicator
and then chooses a community indicator  from
from  . (Each indicator points to one of the K communities that its corresponding node is a member of.) If these indicators point to the same community, then it connects nodes i and j with high probability; otherwise, they are likely to be unconnected.
. (Each indicator points to one of the K communities that its corresponding node is a member of.) If these indicators point to the same community, then it connects nodes i and j with high probability; otherwise, they are likely to be unconnected.
These assumptions capture that the connections between nodes can be explained by their memberships in multiple communities, even if we do not know where those communities lie. To see this, we consider a single pair of nodes 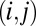 and compute the probability that the model connects them, conditional on their community memberships. This computation requires that we marginalize out the value of the latent indicators
and compute the probability that the model connects them, conditional on their community memberships. This computation requires that we marginalize out the value of the latent indicators  and
and  .
.
Let  be the probability that two nodes are connected given that their community indicators are both equal to k. For now, assume that if the indicators point to different communities then the two nodes have zero probability of being connected. (In the full model, they will also have a small probability of being connected when the indicators are different, but this simplified version gives the intuition.) The conditional probability of a connection is as follows:
be the probability that two nodes are connected given that their community indicators are both equal to k. For now, assume that if the indicators point to different communities then the two nodes have zero probability of being connected. (In the full model, they will also have a small probability of being connected when the indicators are different, but this simplified version gives the intuition.) The conditional probability of a connection is as follows:

The first two terms represent the probability that both nodes draw an indicator for the kth community from their memberships; the last term represents the conditional probability that they are connected given that they both drew that indicator. (The parameter 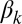 relates to how densely connected the kth community is.) The probability that nodes i and j are connected will be high when
relates to how densely connected the kth community is.) The probability that nodes i and j are connected will be high when  and
and 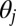 share high weight for at least one community, such as if the social network members attended the same school; it will be low if there is little overlap in their communities. The summation marginalizes out the communities, capturing that the model is indifferent to which communities the nodes have in common. The model captures assortativity—nodes with similar memberships will more likely link to each other (28, 29).
share high weight for at least one community, such as if the social network members attended the same school; it will be low if there is little overlap in their communities. The summation marginalizes out the communities, capturing that the model is indifferent to which communities the nodes have in common. The model captures assortativity—nodes with similar memberships will more likely link to each other (28, 29).
We described the probability that governs a single connection between a pair of nodes. For the full network, the model assumes the following generative process:
1. For each node, draw community memberships
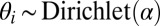 .
.2. For each pair of nodes i and j, where
 :
:(a) Draw community indicator

(b) Draw community indicator

(c) Draw the connection between them from

This defines a joint probability distribution over the N per-node community memberships θ, the per-pair community indicators z, and the observed network y (both links and nonlinks). We will use this as a model of an undirected graph, but it easily generalizes to the directed case.
Given an observed network, the model defines a posterior distribution—the conditional distribution of the hidden community structure—that gives a decomposition of the nodes into K overlapping communities. In particular, the posterior will place higher probability on configurations of the community memberships that describe densely connected communities. With this posterior, we can investigate the set of communities each node participates in and which specific communities are responsible for each of the observed links. In this sense, our algorithm discovers link communities (2, 9). Our visualization in Fig. 1 illustrates this posterior superimposed on a subgraph of the original network.
As for many interesting Bayesian models, however, this posterior is intractable to compute. Furthermore, existing approximation methods like Markov chain Monte Carlo (30) or variational inference (24) are inefficient for real-world–sized networks because they must iteratively consider all pairs of nodes (5, 6, 10). In the next section, we develop an efficient algorithm for approximating the posterior with massive networks.
Finally, we note an exception. The Poisson community model (2) is a probabilistic model of overlapping communities that avoids the all-pairs computation and can be efficiently estimated. We show below that the Poisson model can be good for uncovering true community structure. But we also found that it cannot compute held-out probabilities of links, which makes it ineffective for prediction or for model metrics based on prediction (e.g., to select K).
Posterior Inference.
Our modeling assumptions capture the intuition that each node belongs to multiple communities. To examine observed networks under these assumptions, we compute the posterior distribution of the community structure,

This posterior cannot be computed exactly. (In practice, the community densities βk are also hidden variables. For simplicity we treat them here as fixed parameters but give all details in SI Text.)
The numerator is easy to compute. It is the joint distribution defined by the modeling assumptions. The problem is with the denominator. It is the marginal probability of the data, which implicitly sums over all possible hidden community structures,
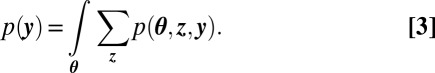
Computing the marginal requires a complicated integral over N simplicial variables and a summation over the  configurations of community indicators. This is exacerbated by the number of nodes N being large. Thus, we approximate the posterior.
configurations of community indicators. This is exacerbated by the number of nodes N being large. Thus, we approximate the posterior.
As we described above, our algorithm to approximate the posterior iterates between subsampling the network, analyzing the subsample, and updating the estimated community structure. This computational structure lets us approximate the posterior with massive networks. It emerges when we adapt two key ideas to the problem: mean-field variational inference and stochastic optimization.
Mean-Field Variational Inference.
Variational inference is a powerful approach to approximate posterior inference in complex probabilistic models (24). It has been adapted to a variety of probabilistic models, although its roots are in the statistical physics literature (31). Variational inference algorithms approximate the posterior in Eq. 2 by defining a parameterized family of distributions over the hidden variables and then fitting the parameters to find a distribution that is close to the posterior. Closeness is measured with Kullback–Leibler (KL) divergence (32). Thus, the problem of posterior inference becomes an optimization problem.
The mean-field variational family independently considers each hidden variable with a different parameterized distribution. In our model, the mean-field variational family is as follows:
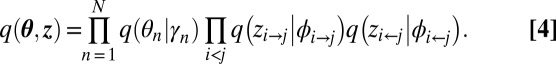
Each factor is in the same family as the corresponding component in the model, but there is a different independent distribution for each instance of each hidden variable.
For example, the model contains a single Dirichlet distribution that specifies the prior over community memberships 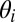 . However, the variational distribution
. However, the variational distribution 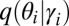 has a different Dirichlet distribution for each node. Once fit, the variational parameter
has a different Dirichlet distribution for each node. Once fit, the variational parameter  captures the posterior distribution of the nth node’s community memberships. Similarly, the model defines the distribution of community indicators based on the corresponding nodes’ community memberships. However, the variational distributions for those indicators are freely parameterized discrete distributions. Once fit, the parameters
captures the posterior distribution of the nth node’s community memberships. Similarly, the model defines the distribution of community indicators based on the corresponding nodes’ community memberships. However, the variational distributions for those indicators are freely parameterized discrete distributions. Once fit, the parameters  and
and  describe discrete distributions that capture the posterior distribution of the indicators when considering the possible connection between i and j. Each variable having its own distribution makes the variational family very flexible. With N nodes, it can uniquely describe each node’s community memberships and which community is activated for each pair of nodes’ possible connection.
describe discrete distributions that capture the posterior distribution of the indicators when considering the possible connection between i and j. Each variable having its own distribution makes the variational family very flexible. With N nodes, it can uniquely describe each node’s community memberships and which community is activated for each pair of nodes’ possible connection.
The variational family  reflects the hidden structure of the observed network when we optimize the variational parameters to minimize the KL divergence to the posterior. Thus, the variational problem is to solve the following:
reflects the hidden structure of the observed network when we optimize the variational parameters to minimize the KL divergence to the posterior. Thus, the variational problem is to solve the following:

We then use the optimal  as a proxy for the true posterior, for example to identify communities in the data or to make predictions about as-yet-unseen links. The optimization connects the variational distribution to the data.
as a proxy for the true posterior, for example to identify communities in the data or to make predictions about as-yet-unseen links. The optimization connects the variational distribution to the data.
Unfortunately, we cannot minimize the KL divergence directly—it is difficult to compute for the same reason that the posterior is difficult to compute—but we can optimize an objective function that is equal to the negative KL divergence up to a constant. (Its optimum is the same as the optimum of the KL objective.) Let  be the entropy of a distribution. The variational objective is
be the entropy of a distribution. The variational objective is

where the expectation is taken with respect to the variational distribution. This objective is a function of the variational parameters in Eq. 4. It relates to the free energy in statistical physics; the first term is the internal energy, with the temperature set to 1.
Maximizing this objective is equivalent to minimizing the KL divergence to the posterior. Intuitively, the first term captures how well  describes a distribution that is likely under the model, keeping both the priors and data in mind through the joint distribution; the second term encourages the variational distribution to be entropic, i.e., this protects it from “overfitting.” Traditional variational inference optimizes the objective with coordinate ascent, iteratively optimizing each variational parameter while holding the others fixed. This has been a successful approach for probability models of small networks (6, 10).
describes a distribution that is likely under the model, keeping both the priors and data in mind through the joint distribution; the second term encourages the variational distribution to be entropic, i.e., this protects it from “overfitting.” Traditional variational inference optimizes the objective with coordinate ascent, iteratively optimizing each variational parameter while holding the others fixed. This has been a successful approach for probability models of small networks (6, 10).
However, traditional variational methods for overlapping community detection do not scale well to real-world–sized networks; previous work has only analyzed networks in the hundreds of nodes. The problem is that there are  terms in the objective function and
terms in the objective function and 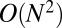 variational parameters. Coordinate ascent inference must consider each pair of nodes at each iteration (10), but even a single pass through a large network can be prohibitive. Our variational inference algorithm avoids this issue through stochastic optimization.
variational parameters. Coordinate ascent inference must consider each pair of nodes at each iteration (10), but even a single pass through a large network can be prohibitive. Our variational inference algorithm avoids this issue through stochastic optimization.
Stochastic Optimization of the Variational Objective.
Stochastic optimization algorithms follow noisy estimates of the gradient of an objective with a decreasing step size. In their classic paper, Robbins and Monro showed that with certain step-size schedules, such algorithms provably lead to the optimum of a convex function (25). (In our case, they provably lead to a local optimum.) Since the 1950s, stochastic optimization has blossomed into a field of its own (33).
Stochastic optimization is particularly efficient when the objective is a sum of terms, as is the case for the variational objective of our model. In these settings, we cheaply compute a stochastic gradient by first subsampling a subset of terms and then forming an appropriately scaled gradient. The scaled gradient is a random variable whose expectation is the true gradient.
Specifically, the variational objective for our model contains a sum of terms for each pair of nodes. Thus, our algorithm iteratively subsamples a subset of pairs and then updates its current estimate of the community structure by following a scaled gradient computed only on that subset. (By “community structure,” we mean the  parameters γ, which describe the posterior distribution of each node’s community memberships θ.) This is a form of stochastic variational inference algorithm (26). At iteration t, we
parameters γ, which describe the posterior distribution of each node’s community memberships θ.) This is a form of stochastic variational inference algorithm (26). At iteration t, we
Subsample a set of pairs of nodes
 .
.For each pair
 , use the current community structure to compute the indicator parameters
, use the current community structure to compute the indicator parameters  and
and 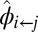 .
.Adjust the community memberships γ.
Repeat.
The details of how we find the optimal indicator parameters and how we adjust the community memberships are in SI Text. What is important about our algorithm is that it does not require analyzing all  pairs at each iteration and that it is a valid stochastic optimization algorithm of the variational objective. It scales to massive networks.
pairs at each iteration and that it is a valid stochastic optimization algorithm of the variational objective. It scales to massive networks.
Our algorithm is flexible in terms of how we sample the subset of pairs in step 1. We can analyze all of the pairs associated with a sampled node; or we can use a subsampling technique that makes data collection easier, for example if the network is stored in a distributed way. We have explored several methods of subsampling pairs of nodes:
Sample uniformly from the set of all pairs.
Sample a node and select its linked pairs.
Sample a node and select all its pairs (links and nonlinks).
Naively applied, biased sampling strategies lead to biases in approximate posterior inference. In SI Text, we show how to correct for these biases. Using these strategies can lead to faster convergence of the variational distribution (34).
We emphasize that our algorithm does not prune the network to make computation manageable (18). Rather, it repeatedly subsamples subgraphs at each iteration. Furthermore, we do not need to have collected the entire network to run the algorithm. Because it operates on subsamples, it gives a natural approach for interleaving data collection and model estimation.
The Number of Communities.
Probabilistic models of community detection require setting the number of communities, and in typical applications we will want to set this number based on the data. In our empirical study, we addressed this model selection problem in two ways. One was by evaluating the predictive performance of the model for varying numbers of communities. A second way was to set the number of communities as part of the initialization procedure of the variational distribution. This is detailed in SI Text. It works well and is faster than the predictive approach.
A Study of Real and Synthetic Networks
We studied our algorithm on real and synthetic networks. With real networks, we demonstrate how it can help us explore massive data: on networks with millions of nodes, it identifies overlapping communities and the nodes that bridge them. On synthetic data, where the ground truth is known, we confirm that it accurately identifies the overlapping communities.
Exploring Real-World Network Data.
We first show how our algorithm can be used to study massive real-world networks. We analyzed two citation networks: a network of 575,000 scientific articles from the arXiv preprint server (21) and a network of 3,700,000 patents from the US patent network (35). In these networks, a link indicates that one document cites another. We also analyzed a large network of 875,000 Web pages from Google (36). (These data did not contain the descriptions of the nodes that are required to visualize the communities. Our quantitative analyses of this network are in SI Text.) In all networks, we treated the directed links as undirected—the presence of a link is evidence of similarity between the nodes and is independent of direction. [This is common in hyperlink graph analysis (1).] These networks are much larger than what can easily be analyzed with previous approaches to computing with mixed-membership stochastic blockmodels (10). [Although we note that several efficient methods have recently been developed for blockmodels without overlapping communities (14–16).]
We analyze a network by setting the number of communities K and running the stochastic inference algorithm. (Our software is available at https://github.com/premgopalan/svinet. More details about these fits are in SI Text.) This results in posterior estimates of the community memberships for each node and posterior estimates of the community assignments for each node pair (i.e., for each pair of nodes, estimates of which communities governed whether they are connected). With these estimates, we visualize the network according to the discovered communities.
Scientific Articles from arXiv.
The arXiv network (21) contains scientific articles and citations between them. Our large subset of the arXiv contains 575,000 physics papers. We ran stochastic inference to discover 200 communities.
Fig. 1 illustrates a subgraph of the arXiv network and demonstrates the structure that our algorithm uncovered. In the model, each node i contains community memberships 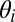 and each link
and each link  is assigned to one of the K communities. In the figure, we colored each link according to the peak of the approximate posterior
is assigned to one of the K communities. In the figure, we colored each link according to the peak of the approximate posterior  . This suggests within which communities and to what degree each paper has had an impact. (We note that most of the links attached to highly cited articles are incoming links, so visualizing these links reveals the communities influenced by the paper.)
. This suggests within which communities and to what degree each paper has had an impact. (We note that most of the links attached to highly cited articles are incoming links, so visualizing these links reveals the communities influenced by the paper.)
The central article in Fig. 1 is the highly cited article “An alternative to compactification” (22), which was published in 1999. The article proposes a simple explanation to one of the most important problems in physics: Why is the weak force 1032 times stronger than gravity? The paper’s external tag (given by the authors) suggests it is primarily a theoretical paper. It has had, however, an impact on a diverse array of problems including certain astrophysics puzzles regarding the structure of the universe (37) and the confrontation between general relativity and experiment (38).
In analyzing the full network of citations, our algorithm has captured how this article has played a role in multiple subfields. It assigned it to membership in nine communities and gave it a high posterior bridgeness score (39), a measure of how strongly it bridges multiple communities. We note that bridgeness is a function of known community memberships. In our networks, the communities are not observed. Thus, we estimated the posterior using our algorithm and then computed the expected bridgeness.
In the subgraph of Fig. 1, the link colors correspond to the research communities associated with the links. We visualize the four top communities that link to this article: “High Energy Physics: Theory,” “High Energy Physics: Phenomenology,” “General Relativity and Quantum Cosmology,” and “Astrophysics.” (Naming and interpreting communities is a difficult problem in unsupervised community detection. For visual convenience, we examine the external tags given to the articles and name each community by its most common tag. Note the algorithm does not have access to the tags.) We emphasize that the citations alone cannot reveal the role of an article in its citation graph—we executed this analysis by first discovering the communities with our algorithm and then using those discovered communities to compute quantities, like bridgeness (39) and link color, that require community assignments.
As an example of a different kind of article, consider “Cosmological constant—the weight of the vacuum” (40). This article has 1,117 citations in the dataset, on the same order as ref. 22. It discusses the theoretical and cosmological aspects of the cosmological constant. Our algorithm finds that this article has a lower bridgeness, and membership in only two communities. Both communities are dominated by the “Astrophysics” subject tag, with the other significant tag being “General Relativity and Quantum Cosmology.” Detecting these two kinds of articles highlights an advantage of this type of analysis. By discovering the hidden community structure, we can separate articles (of similar citation count) that have had interdisciplinary impact from those with impact within their particular fields.
We have illustrated a small subgraph of this large network, centered around a specific article. Across the whole network, we can use the posterior bridgeness to filter and find a collection of articles that have had interdisciplinary impact. In SI Text we show the top 10 papers in the arXiv network by posterior bridgeness. The top scientific articles in the arXiv network have a wide impact, as they concern data, parameters, or theory applied in various subfields of physics. For example, the top article, “Maps of dust infrared emission for use in estimation of reddening and cosmic microwave background radiation foregrounds,” (41) constructs an accurate full sky map of the dust temperature useful in the estimation of cosmic microwave background radiation. This filtering demonstrates the practical potential for unsupervised analysis of large networks. The posterior bridgeness score, a function of the discovered communities, helps us focus on a class of nodes that is otherwise difficult to find.
US Patents.
The National Bureau of Economic Research maintains a large dataset of US patents (35). It contains 3,700,000 patents granted between 1975 and 1999 and the citations between them. We analyzed this network, setting the number of communities to 1,000.
Fig. 2 illustrates a subgraph of the patents data that reveals overlapping community structure around “Process for producing porous products” (42). This patent was issued in 1976 and describes an efficient process for producing highly porous materials from tetrafluoroethylene polymers. It has influenced the design of many everyday materials, such as waterproof laminate, adhesives, printed circuit boards, insulated conductors, dental floss, and strings of musical instruments. Our algorithm assigned it a high posterior bridgeness and membership in 39 communities. The classification tags of the citing patents confirm that it has influenced several areas of patents: Synthetic Resins or Natural Rubbers, Prosthesis, Stock Material, Plastic and Nonmetallic Article Shaping, Adhesive bonding, Conductors and Insulators, and Web or Sheet. Fig. 2 illustrates the top communities for this patent, found by our algorithm.
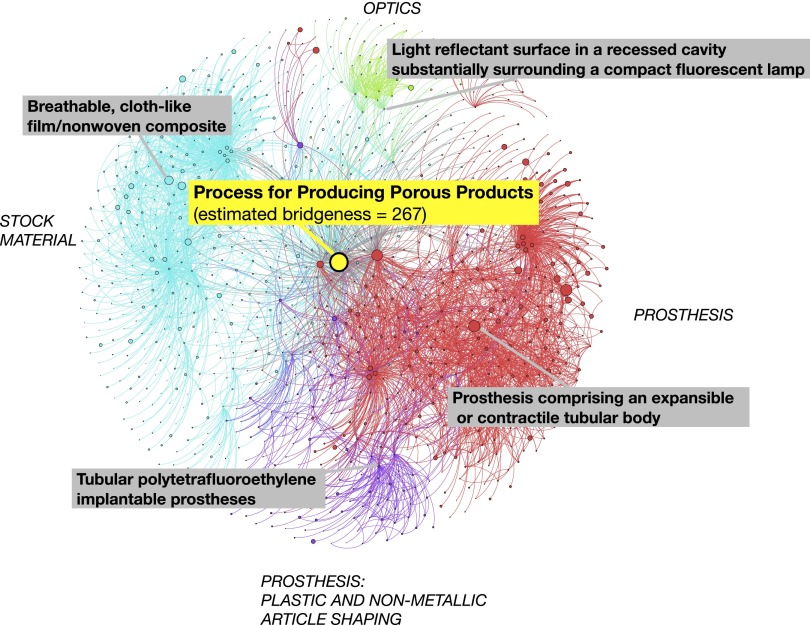
The discovered community structure in a subgraph of the US Patents network (35). The figure shows subgraphs of the top four communities that include citations to “Process for producing porous products” (42). We visualize the links between the patents and show titles of some of the highly cited patents. Each community is labeled with its dominant classification; nodes are sized by their bridgeness (39); the local network is visualized using the Fruchterman–Reingold algorithm (46). This is taken from an analysis of the full 3.7 million node network.
We also studied a patent with a comparable number of citations but with significantly lower bridgeness. “Self-controlled release device for administering beneficial agent to recipient” (43) concerns a novel osmotic dispenser for continually administering agents, e.g., ophthalmic drugs. It has 339 citations, comparable to the 441 of ref. 42, but a much lower bridgeness score. Our algorithm assigned it to seven communities, with the classification tags mostly restricted to “Drug: Bio-Affecting and Body Treating Compositions” and “Surgery.”
Comparisons to Ground Truth on Synthetic Networks.
We demonstrated that our algorithm can help explore massive real-world networks. As further validation, we performed a benchmark comparison on synthetic networks where the overlapping communities are known. We used the “benchmark” tool (44) to synthesize networks with the number of nodes ranging from one thousand to one million.
We compared our algorithm to the best existing algorithms for detecting overlapping communities (2, 8, 9, 11–13, 17). Each algorithm analyzes the (unlabeled) network and returns both the number of communities and community assignments for each node. In our algorithm, we chose the number of communities in an initialization phase of variational inference. [The details are in SI Text. Note that the Poisson community model (2) also requires setting the number of communities. We used the same number of communities derived from our initialization procedure.] A better algorithm better recovers the true community structure. We measured closeness to the truth with the normalized mutual information (NMI) (44), which measures the strength of the relationship between the true and discovered labels.
Most methods could not scale to one million node networks. The four that did were our algorithm, the Poisson community model (2), COPRA (12), and INFOMAP (13). Fig. 3 shows the NMI for these methods on 15 synthetic networks, 5 each of 10,000 nodes, 100,000 nodes, and 1,000,000 nodes. See SI Text for the full table of results.
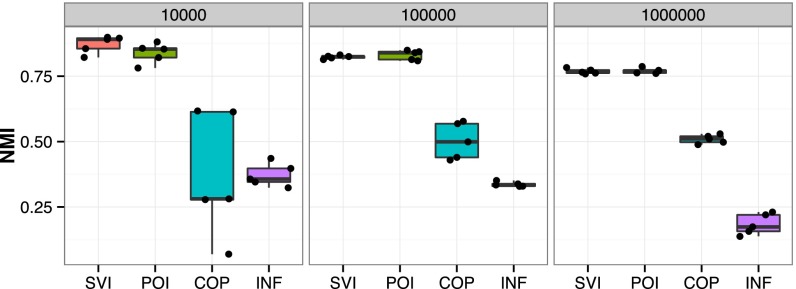
The performance of scalable algorithms on synthetic networks with overlapping communities. The numbers of nodes in each network span ten thousand to one million, and for each network size we generated five networks. Our stochastic inference algorithm (SVI) outperforms scalable alternatives, the INFOMAP algorithm (INF) (13) and the COPRA algorithm (COP) (12), while performing as well as the Poisson community model (POI) (2). We measure accuracy with normalized mutual information (NMI) (44). We also compared with many other methods that could not scale up to one million nodes; see SI Text for a full table of results.
Discussion
We have developed and studied a scalable algorithm for discovering overlapping communities in massive networks. Our approach naturally interleaves subsampling the network and reestimating its community structure. We focused on a specific Bayesian model but we emphasize that this strategy can be used to accommodate many kinds of assumptions. For example, we can posit varying degree distributions to better capture the expected properties of real networks or use Bayesian nonparametric assumptions (45) to infer the number of communities within the analysis. In general, with the ideas presented here, we can use sophisticated statistical models to analyze massive real-world networks.
Acknowledgments
D.M.B. is supported by Office of Naval Research Grant N00014-11-1-0651, National Science Foundation CAREER 0745520, BIGDATA IIS-1247664 and The Alfred P. Sloan Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.H.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/10.1073/pnas.1221839110/-/DCSupplemental.
References
Articles from Proceedings of the National Academy of Sciences of the United States of America are provided here courtesy of National Academy of Sciences
Full text links
Read article at publisher's site: https://doi.org/10.1073/pnas.1221839110
Read article for free, from open access legal sources, via Unpaywall:
https://www.pnas.org/content/pnas/110/36/14534.full.pdf
Citations & impact
Impact metrics
Citations of article over time
Alternative metrics
Smart citations by scite.ai
Explore citation contexts and check if this article has been
supported or disputed.
https://scite.ai/reports/10.1073/pnas.1221839110
Article citations
Multimodal measures of spontaneous brain activity reveal both common and divergent patterns of cortical functional organization.
Nat Commun, 15(1):229, 03 Jan 2024
Cited by: 5 articles | PMID: 38172111 | PMCID: PMC10764905
Living on the edge: network neuroscience beyond nodes.
Trends Cogn Sci, 27(11):1068-1084, 14 Sep 2023
Cited by: 4 articles | PMID: 37716895 | PMCID: PMC10592364
Review Free full text in Europe PMC
LazyFox: fast and parallelized overlapping community detection in large graphs.
PeerJ Comput Sci, 9:e1291, 20 Apr 2023
Cited by: 1 article | PMID: 37346513 | PMCID: PMC10280410
Estimating Mixed Memberships in Directed Networks by Spectral Clustering.
Entropy (Basel), 25(2):345, 13 Feb 2023
Cited by: 0 articles | PMID: 36832711 | PMCID: PMC9955123
A population-based study of precision health assessments using multi-omics network-derived biological functional modules.
Cell Rep Med, 3(12):100847, 08 Dec 2022
Cited by: 2 articles | PMID: 36493776 | PMCID: PMC9798030
Go to all (25) article citations
Data
Data behind the article
This data has been text mined from the article, or deposited into data resources.
BioStudies: supplemental material and supporting data
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Dynamic social community detection and its applications.
PLoS One, 9(4):e91431, 10 Apr 2014
Cited by: 5 articles | PMID: 24722164 | PMCID: PMC3982965
Discovering link communities in complex networks by an integer programming model and a genetic algorithm.
PLoS One, 8(12):e83739, 30 Dec 2013
Cited by: 5 articles | PMID: 24386268 | PMCID: PMC3875478
Efficient and principled method for detecting communities in networks.
Phys Rev E Stat Nonlin Soft Matter Phys, 84(3 pt 2):036103, 08 Sep 2011
Cited by: 32 articles | PMID: 22060452
Community structure in social and biological networks.
Proc Natl Acad Sci U S A, 99(12):7821-7826, 01 Jun 2002
Cited by: 1886 articles | PMID: 12060727 | PMCID: PMC122977
Review Free full text in Europe PMC


